Sympatric Distribution of Three Human Taenia Tapeworms Collected between 1935 and 2005 in Korea
Article information
Abstract
Taeniasis has been known as one of the prevalent parasitic infections in Korea. Until recently, Taenia saginata had long been considered a dominant, and widely distributed species but epidemiological profiles of human Taenia species in Korea still remain unclear. In order to better understand distribution patterns of human Taenia tapeworms in Korea, partial nucleotide sequences of mitochondrial cox1 and ITS2 (internal transcribed spacer 2) were determined, along with morphological examinations, on 68 Taenia specimens obtained from university museum collections deposited since 1935. Genomic DNA was extracted from formalin-preserved specimens. Phylogenetic relationships among the genotypes (cox1 haplotype) detected in this study were inferred using the neighbor-joining method as a tree building method. Morphological and genetic analyses identified 3 specimens as T. solium, 51 specimens as T. asiatica, and 14 specimens as T. saginata. Our results indicate that all 3 Taenia tapeworms are sympatrically distributed in Korea with T. asiatica dominating over T. saginata and T. solium.
INTRODUCTION
The cestodes of medical importance in Korea reportedly include Taenia and Diphyllobothrium tapeworms. Taeniasis and diphyllobothriasis are among the most prevalent types of human cestode infections in Korea [1]. Human taeniasis has become prevalent in Korea since the first report in 1924 [2]. A nationwide study was reported for the first time in 1969 by Seo et al. [3], and revealed a positive rate of 0.7% for taeniid eggs. Subsequent nationwide surveys conducted every 5 yr by MHSA (Ministry of Health and Social Affairs) and KAHP (Korea Association of Health Promotion) have revealed that the positivity rate for taeniid eggs decreased from 1.9% in 1969 to 0.02% in 1997 and human taeniasis was no more detected in the 2004 KAHP's survey. The infection ratio of T. solium versus T. saginata was roughly about 1 : 5 based on the collective data from researches reported between 1965 and 1988 [4]. The prevalence of taeniasis among Koreans was considered to be caused by infection with T. saginata until Eom and Rim [5] revised the identification of the Korean Taenia tapeworm to establish a new species, T. asiatica. Differentiating human-infecting Taenia tapeworms is based mostly on the number of uterine branches in the proglottids or on the absence or presence of hooks on the scolex. However, the specificity of differentiation is low for direct observations of these tapeworms due to their morphological similarities, particularly in the eggs and proglottids of adult worms. Differential species identification between T. saginata and T. asiatica becomes even more difficult when stool sample examination is required because egg morphology is almost indistinguishable under light microcopy.
Epidemiologically, T. asiatica has been found in Taiwan, Korea, China, Indonesia, the Philippines, and Vietnam [4,6-11], and the overlapping distributions of the 2 tapeworms in these countries cause frequent confusion. The epidemiological status of the infections with T. asiatica, T. saginata, and T. solium were questionable in Korea, where this has been studied based on raw-food eating habits [4]. Consequently, it was necessary to clarify the distributions of these tapeworms in Korea.
Molecular approaches to differential diagnosis of human Taenia tapeworms have been developed in recent years, including the use of sequence-specific DNA probes, PCR-RFLP (restriction fragment length polymorphisms) [12,13], and multiplex PCR [15,16]. The present study used differences in the morphology, sequence variations in the mitochondrial cytochrome c oxidase I gene, and rDNA internal transcribed spacer II as markers for identifying 3 human Taenia tapeworms in Korea.
MATERIALS AND METHODS
Specimens
A total of 68 Taenia tapeworms were analyzed in this study (Table 1). These specimens were collected between 1935 and 2005 in the Republic of Korea. Some Taenia tapeworm proglottids were passed naturally in stools, or strobilae were discharged after treatment with niclosamide or praziquantel and purgation. The morphological features of the strobilae and proglottids were compatible with those of Taenia tapeworms. Mature proglottids were examined by light microscopy with carmine staining on the press-and-fixed specimen in an acetic-acid-formalin-alcohol solution. Twenty-nine specimens were preserved in 10% formalin, and 22 were kept in 70% ethanol, and 17 were kept in -20℃ or -70℃.
DNA extraction
Total genomic DNA was extracted from the tissues of terminal proglottids of Taenia tapeworms using the DNeasy Tissue Kit (Qiagen, Valencia, California, USA) according to the manufacturer's instructions and used as the template DNA for PCR amplification of the target gene fragments. Genomic DNA of specimens preserved in formalin was extracted using modified Azuma et al., [16] and Li et al., [17] protocols. The tissues (0.01 g) were crushed in liquid nitrogen and then soaked in TE buffer for 3-5 hr and then digested for 30 min in the DNA extraction buffer (100 mM NaCl, 50 mM EDTA in pH 8.0, 50 mM Tris in pH 8.0, 10% SDS in pH 7.2, and 20 mg/ml proteinase K) at 56℃. Samples in a hexadecyltrimethylammoniumbromide (CTAB)/NaCl solution were incubated for 3 hr at 65℃. The cellular debris was removed and the genomic DNA was extracted using an extraction protocol with phenol-chloroform-isoamyl-alcohol (25: 24: 1, v/v/v). DNA was precipitated in 3 M sodium acetate (pH 5.2) and a 2 times volume of 95% ethanol. The pellet was dissolved in 50 µl of TE buffer (10 mM Tris-HCl and l mM EDTA in pH 8.0). The genomic DNA was treated with RNAse A at 37℃ for 30 min and kept at -20℃ until use. The extraction efficiency was examined using agarose-gel electrophoresis. The optical densities at 260 and 280 nm (OD260 and OD280, respectively) were measured by spectrophotometry.
PCR
The 5.8S-ITS2-28S regions of nuclear ribosome and the mitochondrial cox1 gene were targeted for PCR amplification. PCR reactions were performed in a reaction mixture of 50 µl with 0.01 µg/µl of genomic DNA, 10 × PCR buffer (20 mM Mg+), 10 mM dNTP mixture, 10 pmoles of each primer, and 2.5 U/µl Taq DNA polymerase (High Fidelity PCR system, Roche, Mannheim, Germany). PCRs were performed in a GeneAmp PCR System 9700 (Applied Biosystems, Langen, Germany) and involved 1 cycle of initial denaturation at 94℃for 3 min, followed by 35 cycles of denaturation (94℃ for 1 min), annealing (46℃ for 1 min for cox1; 66℃ for 30 sec for ITS2), and extension (72℃ for 1 min), with a final extension at 72℃ for 10 min. The PCR primers used were 687 (5'-GTTAGTTTCTTTTCCTC-3') and CS250 (5'-ACCCTGGACGGTGGATCACTCGG-3') for ITS2 which yielded a 534-bp product including 5.8S rDNA and 28S rDNA, corresponding to positions 2723-745 and 3347-3362 of the Drosophila melanogaster rDNA sequence (GenBank M21017); and T1F (5'-ATATTTACTTTAGATCATAAGCGG-3') and T1R (5'-ACGAGAAAATATATTAGT CATAAA-3') for cox1 which yielded a 348-bp product. The PCR amplification products were directly sequenced using DNA sequencer (3730XL, model Applied Biosystems) and the Big-Dye Terminator Cycle Sequencing Reaction Kit (version 3.2, Applied Biosystems).
DNA sequencing and analyses
The sequences of cox1 and the ITS2 regions from Taenia tapeworm specimens were aligned using the CLUSTAL X multiple alignment program [18] and the Bioedit programs (version 5.0.6, BIOSOFT, Ferguson, Missouri, USA). The molecular identification of Taenia tapeworm specimens was based on the similarity of nucleotide sequences and phylogenetic relationships with those of T. asiatica (GenBank NC 004826), T. saginata (GenBank AB066495) and T. solium (GenBank NC 004022). Phylogenetic relationships among the Taenia genotypes were inferred using different tree-constructing algorithms, i.e., neighbor-joining (NJ), maximum parsimony (MP), and minimum evolution (ME), using the Mega 3.1 program [19]. NJ analysis was performed based on corrected distance matrix of the Kimura's 2 parameter substitution model. Bootstrap analysis was performed with 3,000 replications. The mitochondrial cox1 gene sequence of Echinococcus multilocularis (NC 000928) was used as an outgroup for phylogenetic analysis of cox1 dataset.
RESULTS
Sequence divergence of cox1 gene and ITS2
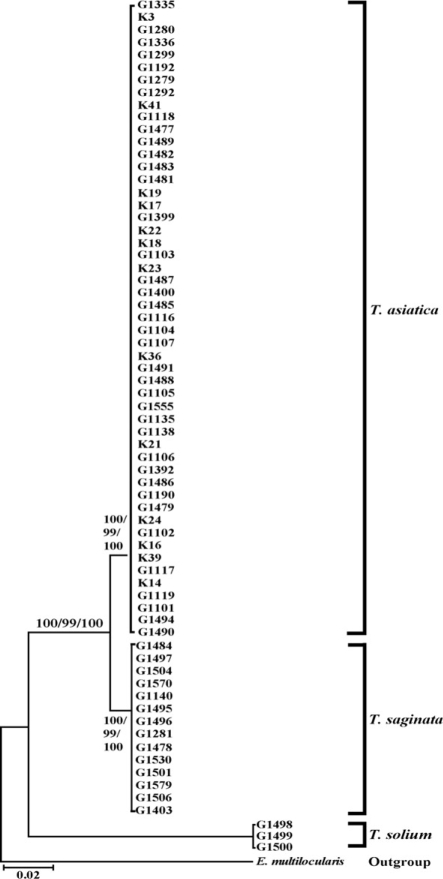
Taenia specimens were molecularly identified by direct sequencing of ITS2 and the partial mitochondrial cox1 region. ITS2 sequences of human Taenia tapeworms were ranged in length from 481 to 512 bp. The various lengths were represented by 503-bp and 512-bp sequences in T. saginata and T. asiatica, respectively, and 481-bp sequences in T. solium. The difference in the number of inferred gap among species ranged between 3 and 36 bp in the human Taenia tapeworms (Fig. 1). The nucleotide differences in these 3 species varied in size from 12 to 31 bp in the ITS2 region. The sequence divergenes of ITS2 between 3 Taenia species ranged from 7.3% (T. saginata and T. asiatica) and 33.7% (T. asiatica and T. solium), respectively. The cox1 sequences (348 bp) showed 59 polymorphic sites, with 28 sites of nonsynonymous substitutions in human Taenia tapeworms (Table 2). The sequences of T. asiatica and T. saginata differed by 7%, while those of T. saginata and T. solium differed by 15%. Phylogenetic analysis of the mtDNA cox1 sequences for a total of 68 isolates (of which 51 were obtained from 9 identified provinces in Korea) identified T. solium as basal to the T. saginata-T. asiatica clade. Trees topology using various analytic methods (NJ, MP, and ME) generated was identical with very high confidence values (bootstrap values of 100%, 99%, and 100% in NJ, MP, and ME, respectively) for 3 major branches representing each of 3 species (Fig. 2).

Sequence alignment of ITS2 regions of human Taenia tapeworms. Dot (.) indicates identical nucleotide sequence to T. asiatica. Gap (-) was introduced for maximum alignments.
Species composition and spatial distribution
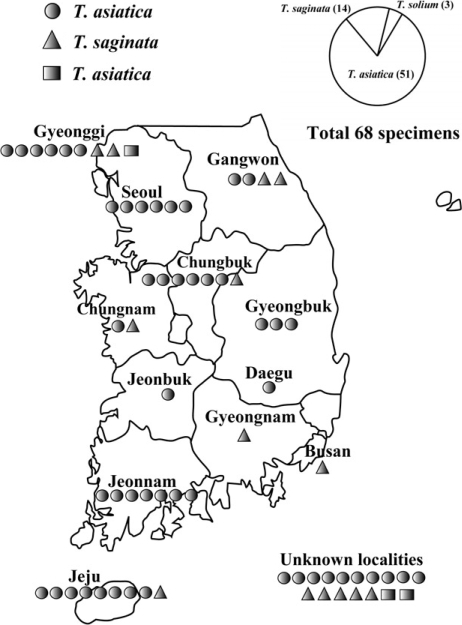
Morphological and genetic analyses identified 3 specimens (from Gyeonggi and unknown localities) as Taenia solium, 51 specimens (from Chungbuk, Gangwon, Gyeonggi, Gyeongbuk, Jeonnam, Jeonbuk, Chungnam, Jeju, Seoul, Daegu, and unknown localities) as T. asiatica and 14 specimens (from Chungbuk, Gangwon, Gyeonggi, Chungnam, Busan, and unknown localities) as T. saginata. Most provinces exhibited 1 or 2 kinds of tapeworms except for Gyeonggi province that showed all 3 species of tapeworms. Ratios of T. solium: T. asiatica: T. saginata calculated from both morphological and molecular data was 3: 51: 14 (or approximately 1: 17: 5). T. solium, T. saginata, and T. asiatica tapeworm infections were only exhibited in Gyeonggi province. T. saginata and T. asiatica tapeworm infections were presented in Gangwon, Chungbuk, Chungnam and Jeju provinces (Fig. 3). This result indicates that T. asiatica was the dominant human-infecting Taenia tapeworm in Korea.
DISCUSSION
Two types of taeniases (T. saginata and T. solium) were only recognized in Korea until 1992, with T. asiatica being newly described in 1993 as the third type of causative agent. T. saginata is morphologically distinct from T. asiatica by unarmed restellum, large number of uterine buds, and a posterior protuberance on the proglottids. Nevertheless, differential diagnosis of these species is often a task of specialists with much experience, especially when this is performed in field research conditions, which is hampered by the shortage of parasite materials obtained for examination or variable fixation/preservation conditions of the adult worms collected. These 2 species also exhibit different life cycles: with T. asiatica uses suids as intermediate hosts, whereas T. saginata utilizes bovids as intermediate hosts in order to complete their life cycles. This criterion, however, is not applicable in the laboratory condition. Even though the epidemiological understanding of T. asiatica infection remained questionable, T. asiatica rather than T. saginata, had been considered to be the predominant species in Koera [4]. It was therefore necessary to quantify the prevalence of this tapeworm as well as those of T. saginata and T. solium. There was no Taenia egg positive case in the last nationwide survey of the presence of helminth eggs among Koreans, implemented in 2004, suggesting that the occurrence of Taneia species is rapidly decreasing. The difficulties of finding out Taenia-tapeworm-infected individuals prompted us to plan to analyze collections available over all time, dating back up to 1935 from laboratories and institutes, and including any sources of tapeworms in Korea. Recent ad- vancements in DNA analysis techniques made it possible to differentiate species precisely, but the results were not consistent with epidemiological finding.
Twenty-nine of the 68 examined specimens were preserved in 10% formalin, which can cause cross-linking between DNA and protein, and also degrade DNA by reducing sugar, and inflicting oxidative and hydrolytic damage. Therefore, the DNA recovery yield is usually lower for formalin-fixed specimens than ethanol-fixed or frozen specimen. In this study, genomic DNA was extracted from the modified protocol of Azuma et al. [16] and Li et al. [17]. We extended the time of incubation up to 24 hr, and SDS and proteinase K were added during the incubation process. Most of the formalin-preserved samples yielded weak or no PCR amplification, but secondary PCR using the PCR products obtained from the first round of PCR amplifications as a template make it possible to use the PCR product for direct sequencing. The longest time period of formalin preservation used for molecular identification was the specimen (labeled as T. saginata by the original collector) that had been isolated, and kept in 10% formalin since 1935 in the Department of Parasitology, Seoul National University. Very interestingly, cox1 sequence analysis of the specimen identified it as T. asiatica. Overall results were 3 T. solium, 51 T. asiatica, and 14 T. saginata. These results clearly indicate that all 3 human Taneia tapeworms are sympatrically distributed in Korea, with T. asaitica infections accounting for three-quarters of the total Taenia infections.
ACKNOWLEDGEMENTS
This work was supported by a research grant from Chungbuk National University in 2008. Parasite materials used in this study were provided by the Parasite Resource Bank of Korea, National Research Center (R21-2005-000-10007-0), the Republic of Korea.