Molecular Characterization of Nippostrongylus brasiliensis (Nematoda: Heligmosomatidae) from Mus musculus in India
Article information
Abstract
Mus musculus (Rodentia: Muridae) has generally been infected with a rodent hookworm Nippostrongylus brasiliensis. In this report, we present morphological and molecular identification of N. brasiliensis by light and scanning electron microscopy and PCR amplification of mitochondrial cytochrome c oxidase subunit 1 (cox1) gene and the protein sequences encoded by cox1 gene, respectively. Despite the use of N. brasiliensis in many biochemistry studies from India, their taxonomic identification was not fully understood, especially at the species level, and no molecular data is available in GenBank from India. Sequence analysis of cox1 gene in this study revealed that the present specimen showed close identity with the same species available in GenBank, confirming that the species is N. brasiliensis. This study represents the first record of molecular identification of N. brasiliensis from India and the protein structure to better understand the comparative phylogenetic characteristics.
INTRODUCTION
Nippostrongylus Lane, 1923, a heligmonellid genus, is commonly parasitic in the digestive tract of murines. Geographically, Nippostrongylus has been known for a wide range throughout the world [1–5]. Nippostrongylus brasiliensis Travassos, 1914, a gastrointestinal nematode, is a cosmopolitan parasite of a commensal mouse Mus musculus. Our present knowledge of identification of N. brasiliensis nematode is still fragmented in India. Till date, no molecular studies have been reported in India. Recently, mitochondrial genes have been successfully employed as a molecular marker for accurate identification of nematodes [6–8]. The mitochondrial cytochrome c oxidase subunit 1 (cox1) gene has been widely used for identification and phylogenetic studies, and enabled the discrimination of closely related species in nematode phyla [7–9]. The mitochondrial cox1 gene (also known as mtCO1) is a key enzyme of aerobic metabolism, which is located in the inner mitochondrial membrane and a major site for regulation of mitochondrial oxidative phosphorylation. Little information is available regarding parasitic nematodes protein structures and their comparison with isolates of same or closely related species.
During a general survey of the nematode fauna of Mus musculus in the Meerut, U.P., India, several nematodes belonging to Nippostrongylus were collected from the gastrointestinal tract. Their examination using light and scanning electron microscopy revealed that these parasites represented the species N. brasiliensis. Moreover, the specimens were also characterized by using molecular approaches. The mitochondrial cox1 gene was sequenced and analyzed in order to molecularly identify and estimate the validity of N. brasiliensis from Indian region. We also summarized the identification and prediction of cox1 protein structures with comparison of isolates for taxonomic identification with a special focus on the structural aspects through bioinformatics approach.
MATERIALS AND METHODS
A total of 20 M. musculus caught from Meerut (29°01′ N, 77°45′ E), U.P., India were examined for parasitic infections after dissection under chloroform or ether anesthesia. Their gastrointestinal tracts were removed and examined under a stereomicroscope. Total 20 male and 5 female nematodes were recovered from the intestine of M. musculus. They were washed in saline (0.6%) and then fixed in 70% ethanol and stored until studied. For the light microscopy study, the nematodes were mounted in glycerin. A light microscope (Motic SMZ-168, Xiamen, China) equipped with digital image analysis system (Motic Image Plus 2.0 for Windows) and drawing attachment was used for line drawings and morphometric analysis. For scanning electron microscopic studies, parasites were fixed in 70% ethanol, dried by critical point-drier, mounted on SEM studs, and finally coated with a thin layer of gold before being examined with a JOEL Neoscope JCM5000 SEM (Nikon Instruments, Melville, New York, USA) at an accelerating voltage of 10 kV. Prepared slides of male and female N. brasiliensis were deposited in the Museum, Department of Zoology, Chaudhary Charan Singh University, Meerut, (U.P.), India, under the voucher no. Nem/2015/01. Measurements are given in Table 1.
For molecular analysis, genomic DNA was extracted using a DNeasyTM Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Eluted DNA was kept at −20°C until further use. The partial mitochondrial cox1 gene was amplified by PCR using the primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HC02198 (5′-TAAACT TCAGGGTGACCAAAAAATCA-3′) [10] with cycling profile described previously [10]. PCR products were checked on ethidium bromide stained 1% TAE buffer gel and purified by the PurelinkTM Quick Gel Extraction and PCR Purification Combo kit (Invitrogen, Carlsbad, California, USA) following the manufacturer’s instruction. Sequencing was carried out by the same primers using an ABI Big Dye Terminator version 3.1 cycle sequencing kit with an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, California, USA). Sequences were aligned using Clustal W [11] and manually adjusted. Using the BLASTn algorithm, the obtained sequence was compared with those available in the NCBI database (National Centre for Biotechnology Information; http://www.ncbi.nlm.nih.gov).
The phylogenetic tree was built using the maximum likelihood (ML) and Bayesian inference (BI) analyses. DNA pairwise distances were calculated using the Kimura 2 parameter model with the MEGA 6 software [12]. For ML analysis, GTR + G + I model was chosen based on the best fitting substitution model using the Akaike Information Criterion in MEGA 6 [12]. The tree topology was tested by using bootstrapping over 1,000 replications. TOPALi 2.5 [13] was used to construct the tree for BI analysis. For BI analysis, substitution model was tested by the Bayesian Information Criterion and GTR+I+G was chosen. BI analysis was run for 1,000,000 generations, sampling every 100th tree and discarding ‘burn in’ first 25% of the sampled tree. Oesophagostomum columbianum (KC715827) was used as an outgroup for analysis.
For study of protein sequence, a primary sequence analysis of the N. brasiliensis isolates was performed using the ProtParam [14]. The cox1 protein secondary structure analysis of the N. brasiliensis isolates was obtained using the program SOPMA [15]. The cox1 protein sequence of N. brasiliensis and most related isolate sequence alignment were generated by ESPript 3.0 [16]. To carry out the cox1 homology search for N. brasiliensis against Protein Data Bank (PDB) was performed by using SWISS-MODEL [17]. The same model was employed to generate the 3D structure of the N. brasiliensis for cox1. The model with high score was validated by the Phyre 2 [18] and I-TESSER [19]. The model was refined by energy minimization using the NAMD package [20] and subjected to quality evaluation. MEMSAT-SVM and MEMPACK in PSIPRED workbench [21] were used for the prediction of transmembrane helices and topology of N. brasiliensis protein sequence. RAMPAGE [22] was used for quantitative protein structure evaluation of N. Brasiliensis, and the Ramachandran plot was utilized for geometric assessment. To evaluate the quality of the model and study the energy of residue–residue interactions using a distance-based pair potential ProSA program [23] was employed. TM-align [24] was used for the superimposition between Indian and USA isolates of N. brasiliensis protein sequences for comparison.
RESULTS
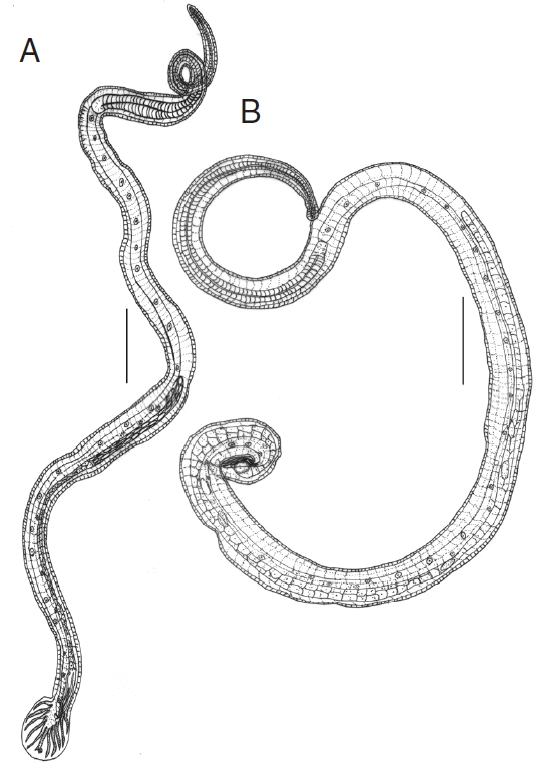
The male and female nematodes collected in this study were diagnosed on the basis of morphological characteristics and preliminarily identified as N. brasiliensis. The male’s bursa with asymmetrical lateral and small dorsal lobes, each branch ending in out of the 3 tips, spicules almost equal, filiform and gubernaculum present (Fig. 1A). Female’s tail conical, pointed, vulva present near anus, and oviparous (Fig. 1B). Measurements were taken and presented in the table (Table 1). SEM images were also provided for the topological view of N. brasiliensis (Fig. 2).
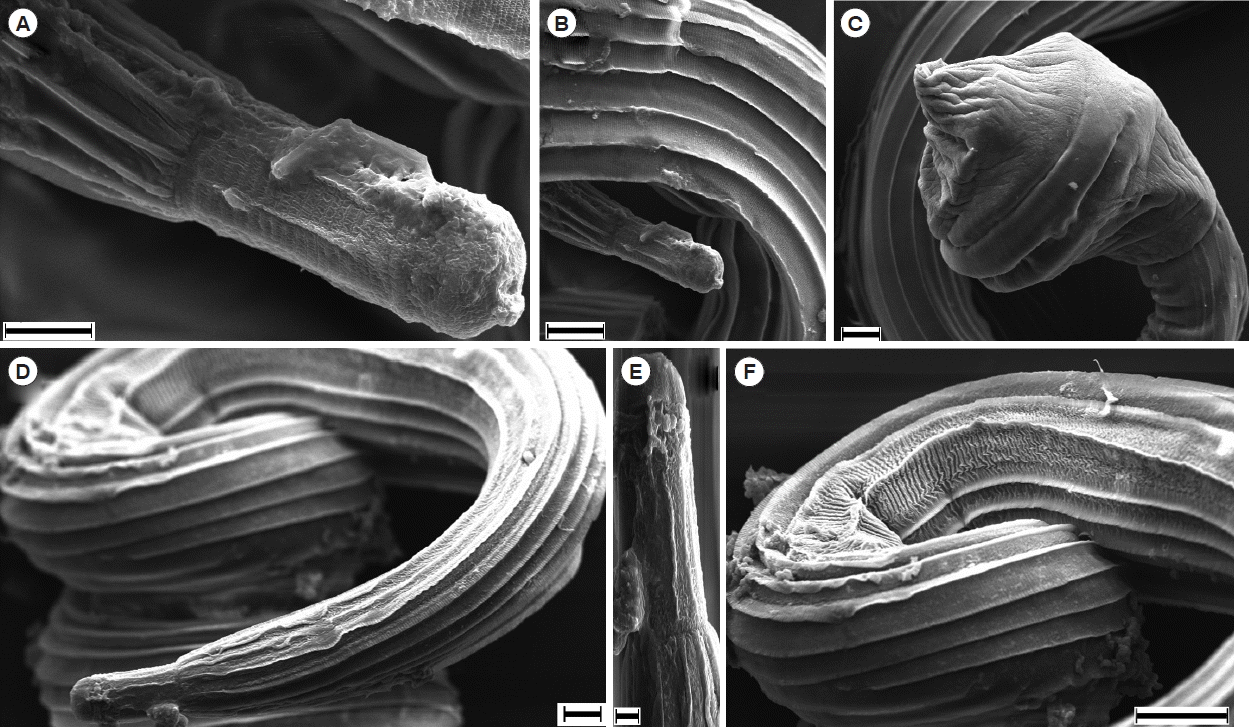
Scanning electron microscopy of N. brasiliensis. (A, B) Anterior portion of male mouth. (C) Tail portion of a male with copulatory bursa. (D, E) Anterior portion of female. (F) Striations on the female body. Scale bars (A) and (D)=10 μm; (B), (C) and (F)=20 μm; (E) 2 μm.
The sequence obtained for the mitochondrial cox1 region was 705 bp in length and deposited in the GenBank database under the accession no. KX146839. There was no N. brasiliensis sequence registered from India till date, thus, comparison was possible with 3 N. brasiliensis sequence (nos. U57035, AF096235, and AF263480) available in GenBank. The available 3 isolates of N. brasiliensis showed a pairwise comparison, 0.14% (U57035), 1.75% (AF096235), and 1.97% (AF263480) nucleotide difference with Indian isolate, respectively. Sequence analysis showed that in all, an isolate from USA (no. U57035) was found the closest to Indian isolate based on the cox1 sequence. Surprisingly, phylogenetic analysis of data showed that 2 isolates of N. brasiliensis (nos. KX146839 and U57035) were located on the same clade, whereas, the other 2 from Korea and UK (nos. AF096235 and AF263480) formed a separate major clade (Fig. 3). This result demonstrated the paraphyly of Nippostrongylus as its isolates were segregated into 2 lineages. According to the phylogenetic tree, N. brasiliensis formed a clade along with Trichostrongylus and Heligmosomoides as the sister group. In addition, all the species that formed the tree belonged to the superfamily Trichostrongyloidea based on mitochondrial cox1 analysis (Fig. 3).
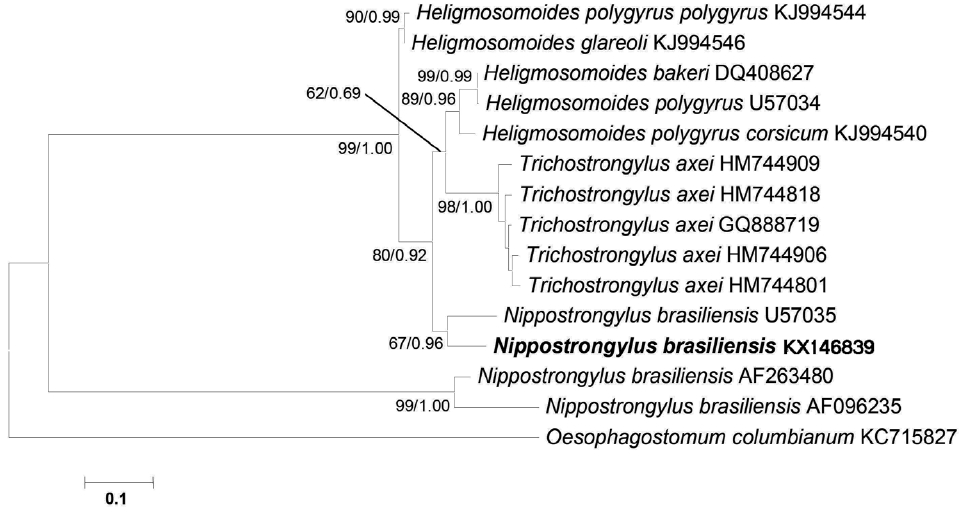
A phylogenetic tree is based on the mitochondrial cox1 sequence demonstrating the position of N. brasiliensis isolate with other nematode species. The tree was generated by maximum likelihood (ML) method. Numbers of nodes indicate bootstrap values (1,000 replications) of ML and posterior probabilities (BI), respectively. GenBank accession nos. are listed along the species names. Species examined in this study are indicated in bold.
Further, we studied the cox1 protein sequence to predict the 3D structure and comparative modeling to show the well conserved structural elements. Results of primary and secondary sequence analysis of cox1 protein sequences of N. brasiliensis isolates available on the NCBI database from various geographical regions are shown in Tables 1 and 2. Sequence alignment and tertiary structure analysis of protein sequence of N. brasiliensis Indian isolate showed the closest similarity with an isolate from USA (Table 3), and template sample (PDB Id: 3abm) against PDB search was presented in Figs. 4 and 5. In Fig. 4A, sequence alignment comparison of the isolate from India and USA with the template sequence is shown. Fig. 4B specifically represents the sequence comparison between Indian and USA isolates of N. brasiliensis with template sequence along with the consensus sequence where except few sites, the nucleotide sequence from Indian isolate showed the highest similarity with USA isolate (Fig. 4C). Fig. 4D depicts about Indian and USA isolate with secondary structure elements on top of the sequences as helices with squiggles, arrows present β-strands, and TT letters show the turns. Below the sequence solvent accessibility is provided by a bar (blue is accessible, cyan is intermediate, white is buried) (Fig. 4D).
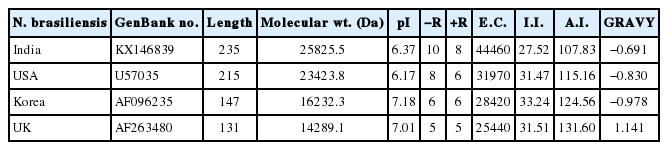
Results of primary sequence analysis of N. brasiliensis isolates of cox1 protein available in NCBI database from various geographical regions
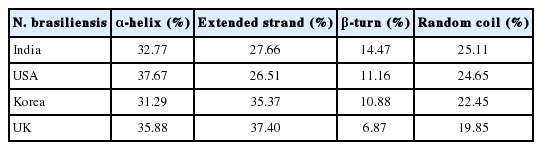
Results of secondary sequence analysis of cox1 data of N. brasiliensis isolates available in the NCBI database from various geographical regions
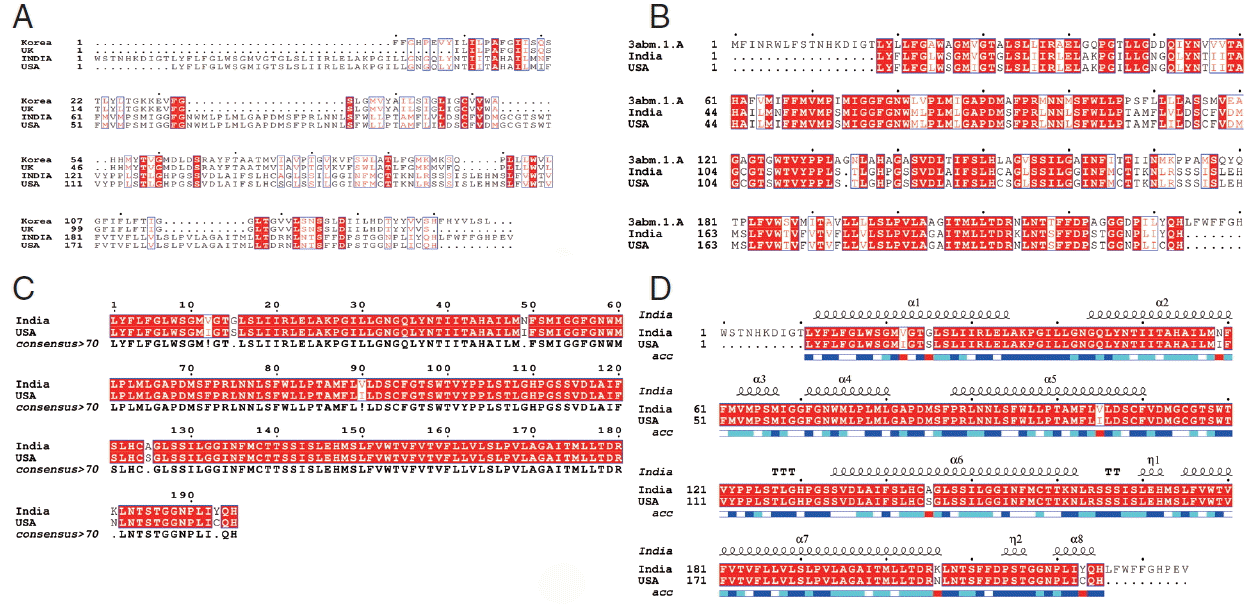
Alignment of the cox1 protein sequence. (A) N. brasiliensis from India and its isolates available on Genbank database from other regions. (B) N. brasiliensis isolate from India and USA along with the template search against the PDB database. (C) N. brasiliensis isolate from India and USA and their consensus sequence (D) showing the N. brasiliensis isolate from India and USA with secondary structure elements presented on top.
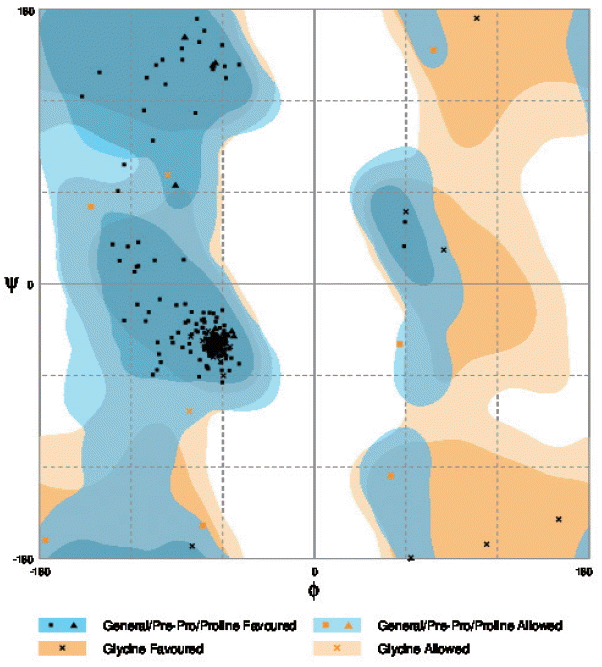
The Ramachandran plot of the structure of N. brasiliensis isolate from India showing residue predicted by RAMPAGE in favored and allowed outer regions.
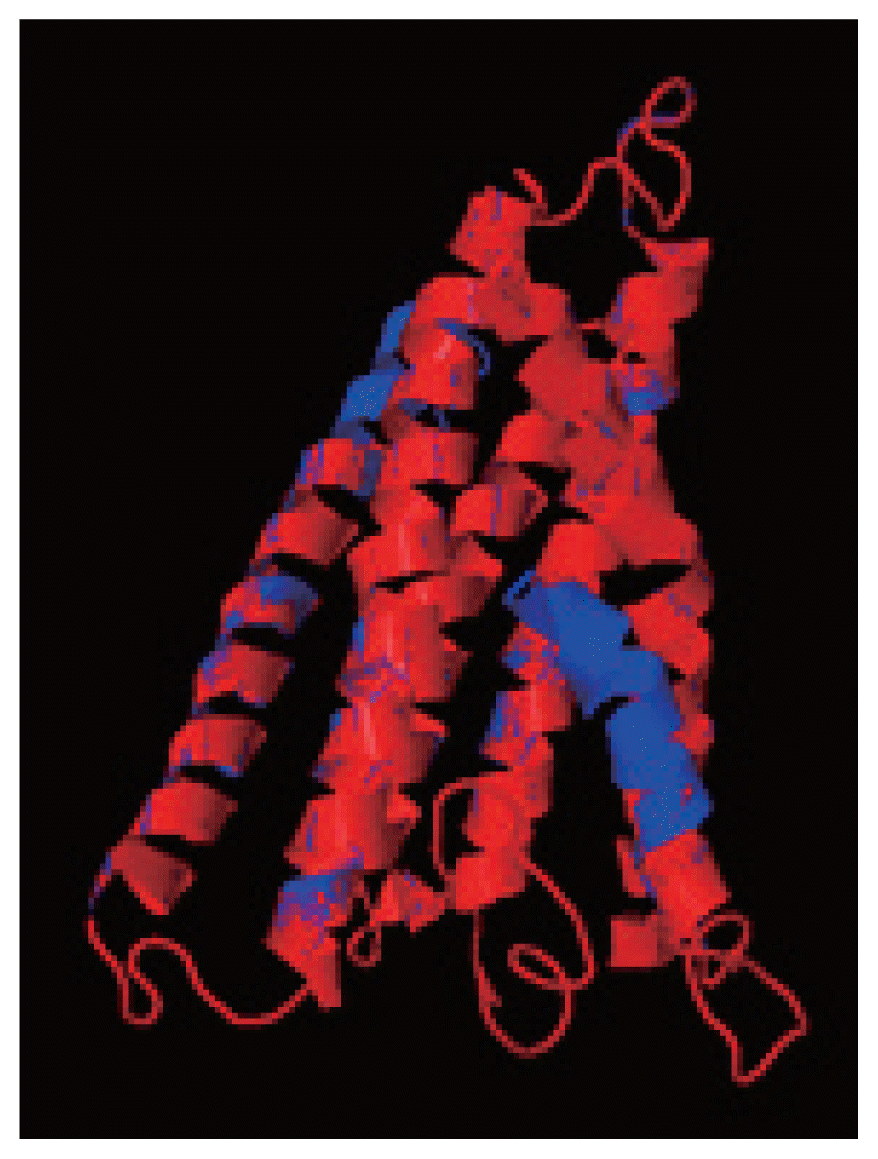
The resulting 3D model of the cox1 protein structure was sorted according to the scores evaluated by protein energy, and the validation of the model was checked by assessing the quality of protein backbone conformation by RAMPAGE for reliability. Rampage φ and ψ regions were used for validation of experimental protein structures. The obtained Ramachandran plot (Psi-Phi) pairs for Indian isolate had 96.6% of residues in most favored regions, while 3.4% residues in generously allowed regions (Fig. 5) whereas for USA isolate was 96.2%. The 3D model of cox1 protein sequence of isolate from India and USA along with sample template (PDB Id: 3abm) were built (Fig. 6). The ProsA analysis showed the overall interaction energy of the model was −3.71 kcal/mol, which is relatively similar to the USA isolate Z score −3.61 kcal/mol. Hydropathy analysis of cox1 protein sequence of isolate from India by MEMSAT-SVM and MEMPACK suggested the presence of 5 transmembrane (TM) helix (Fig. 7). Comparative analysis of cox1 protein structures of isolates from India and USA was performed using TM-align; it compared the 3D structures of proteins and computation structural alignments between 2 protein structures showing the structural similarity. Fig. 8 shows the superimposition of Indian and USA isolates; the 2 isolates were almost similar except few bases that might be due to different geographical regions. The results shown above gave sufficient information of structurally conserved regions in a predicted protein structure.
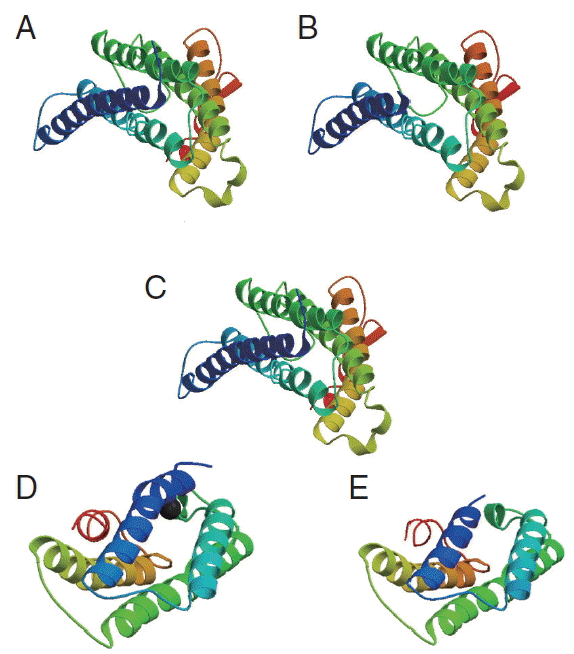
The 3D structure of cox1 showing homology in helices, β-strands, and turns. (A) N. brasiliensis isolate from India. (B) N. brasiliensis isolate from USA. (C) The template (PDB Id: 3abm). (D) N. brasiliensis isolate from Korea. (E) N. brasiliensis isolate from UK.
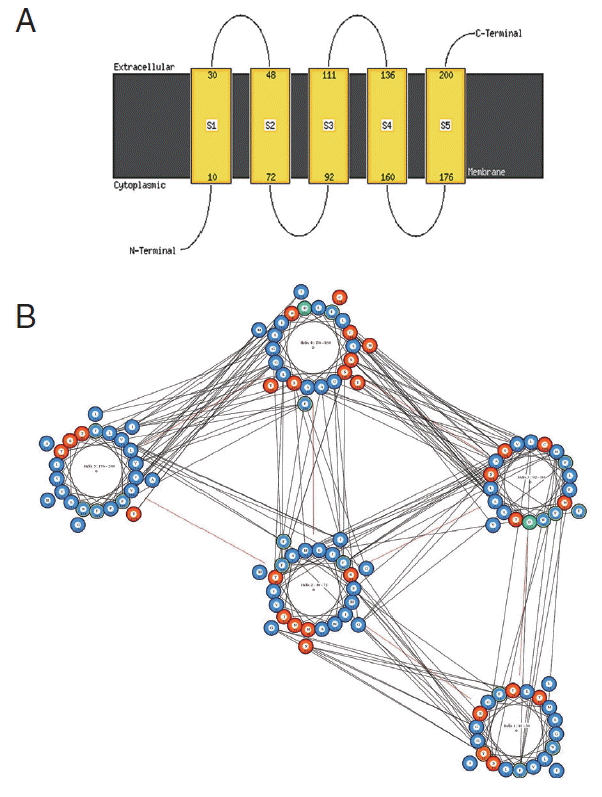
Predicted transmembrane regions of N. brasiliensis isolate from India by (A) MEMSAT-SVM showing TM helices and loop regions and (B) helix orientation cartoon and the predicted helical packing arrangement by MEMPACK, colors in the cartoon indicate hydrophobic residues (blue), polar residues (red), and charged residues (green for negative, purple for positive). The lines between residues indicated a predicted interaction.
DISCUSSION
The present study adds to our knowledge and provides the strong molecular based evidence for the existence of N. brasiliensis from India parasitizing M. musculus as a common parasite of rodents. Male and female parasites were identified based on morphological diagnostic characteristics but molecular data supplemented more confirmation and validation to the study. However, sometimes species identification based only on morphology cannot be considered reliable especially in the cases where species are morphologically indistinguishable. In India, little attention has been paid to molecular studies of nematodes relatively to morphology that represents an obstacle in identification. We report for the first time the use of mitochondrial cox1 gene for molecular identification of N. brasiliensis from India. Mitochondrial DNA evolves very quickly in genomes that is why differences in very closely related species can be easily identified among closely related/within species [25,26]. Phylogenetic analysis clearly stated that in all isolates of N. brasiliensis available on the database had closest homology with the isolate from USA if compared with isolates from Korea and UK as they had very short cox1 protein sequences. All N. brasiliensis isolates did not form a single clade and showed 2 lineages, this might be due to the paraphyly of this genus as also reported earlier, so the results were consistent with these studies [27–29]. Mitochondrial cox1 proteins have been relatively consistent functional and structural contexts over evolutionary time [30] and have been well conserved in their structures as also proved in the results of this study for determining specific relationships. The protein of isolates from India and USA confirmed 5 transmembrane helixes at the same position indicating structural similarity. The outcome of RAMPAGE showed the reliability, and strong acceptance of the predicted structures of both isolates confirmed their structural similarities and the validation of Indian isolate.
In India, there is a need of attempts to integrate molecular analyses as well as morphology that can provide more effective means of characterization for nematodes. The present study added important details and indicated that the analysis of protein structural components was an important technique for obtaining the 3D structure that will help in studying phylogeny and identification of closely related isolates. The present research offered a backbone to understand the functional and structural insight of cox1 protein of N. brasiliensis that can be used for future molecular studies.
ACKNOWLEDGMENTS
The authors would like to thank the Head of the Department of Zoology, Chaudhary Charan Singh University, Meerut (U.P.), India, for laboratory facilities. Special thanks to Prof. Jerzy M. Behnke, School of Biology, University of Nottingham, NG7 2RD, UK, for his help and providing literature for identification of parasite species (N. brasiliensis).
Notes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.