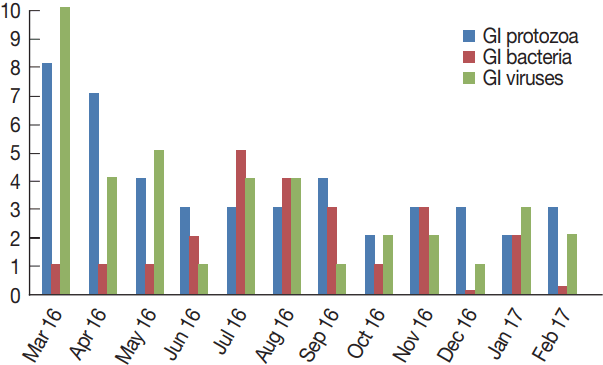High Frequency of Enteric Protozoan, Viral, and Bacterial Potential Pathogens in Community-Acquired Acute Diarrheal Episodes: Evidence Based on Results of Luminex Gastrointestinal Pathogen Panel Assay
Article information
Abstract
Infectious diarrhea is endemic in most developing countries. We aimed to investigate the protozoan, viral, and bacterial causes of acute diarrhea in Taif, Saudi Arabia. A cross-sectional prospective 1-year study was conducted on 163 diarrheal patients of various ages. Stool samples were collected, 1 per patient, and tested for 3 protozoa, 3 viruses, and 9 bacteria with the Luminex Gastrointestinal Pathogen Panel. Overall, 53.4% (87/163) of samples were positives (20.8% protozoa, 19.6% viruses, 2.8% bacteria, and 9.8% mixed). Rotavirus (19.6%), Giardia duodenalis (16.5%), and Cryptosporidium spp. (8.5%) were the mostly detected pathogens. Adenovirus 40/41 (4.2%), Salmonella (3%), Shiga toxin-producing Escherichia coli (3%), and Entamoeba histolytica (2.4%) were also detected. Norovirus GI/II, Vibrio cholerae, Yersinia enterocolitica, and Clostridium difficile toxin A/B were not detected in any patients. All pathogens were involved in coinfections except E. histolytica. Giardia (5.5%) and rotavirus (3%) were the most commonly detected in co-infections. Enterotoxigenic E. coli (2.4%), Campylobacter spp. (2.4%), E. coli 0157 (1.8%), and Shigella spp. (1.2%) were detected in patients only as co-infections. Infections were more in children 0–4 years, less in adults <40 years, and least >40 years, with statistically significant differences in risk across age groups observed with rotavirus (P<0.001), Giardia (P=0.006), and Cryptosporidium (P=0.036) infections. Lastly, infections were not significantly more in the spring. This report demonstrates the high burden of various enteropathogens in the setting. Further studies are needed to define the impact of these findings on the clinical course of the disease.
INTRODUCTION
Infectious diarrhea continues to be a worldwide public health problem. According to an estimate, around 2 billion episodes globally occur per year [1]. Diarrhea is a leading cause of malnutrition and a second leading cause of death in children below the age of 5, killed 5.8 million children in 2015 [2]. Diarrhea can be attributed to a variety of different gastrointestinal (GI) pathogens, including protozoa, viruses, and bacteria. The etiologic diagnosis of infectious diarrhea can be challenging due to 2 reasons. First, the clinical pictures of several infections are often similar, and making diagnosis, based on signs and symptoms, is difficult for physicians. Second, infections can be transmitted to humans via food or water, by person-to-person contact, exposure to animals, or acquired from the environment. Therefore, infection with multiple pathogen species could easily occur [3]. In this case, detection of the concomitant pathogen or the secondary infection is of particular health importance. It could guide physicians to prescribe the most appropriate pathogen-specific medication and health authorities to prevent spread of infections.
Thus, prompt diagnosis of diarrhea should rely on laboratory tests that are subsequently performed based on a presumed etiology by the treating physicians. Nevertheless, the causative pathogen has been unrecognized in about 80% of cases [4]. Indeed, this could be unfavorable for patient management and infection control. Rapid and accurate detection of the suspected GI pathogens in fecal specimens from patients with diarrhea can be also challenging for the microbiology laboratory. It often requires combinations of multiple diagnostic tests, including bacterial culture, antigen detection assay, in-house nucleic acid-based tests, and microscopy. These standard diagnostic test methods can be time-consuming, labor intensive, and exhibits distinct clinical performances [5].
A few years ago, several molecular assays were developed, properly validated, received FDA-clearance, and had a growing popularity for the detection of multiple GI pathogens in human feces [6–9]. The Luminex® xTAG® Gastrointestinal Pathogen Panel (GPP) (Luminex Molecular Diagnostics, Toronto, Canada) was one of these assays. The xTAG GPP was designed to detect or rule out up to 15 pathogens causing diarrhea, in parallel, in 1 stool sample. The sensitivity of the assay has been reported to be higher than that of the conventional laboratory methods by 2 folds or more, according to earlier studies [10,11].
The microbiological etiology of diarrhea has been received much concern among researchers in many parts of the world, including Saudi Arabia [12–14]. However, most of the prior research has been focused on investigating a small range of pathogen species based on the combined use of various diagnostic methods.
In a cross-sectional study, we investigated a range of relevant enteric protozoan, viral, and bacterial pathogens in a population with community-acquired acute diarrheal episodes from Taif Province, Saudi Arabia using the xTAG GPP assay. Our main objective was to estimate the frequency of acute infectious diarrhea caused either by 1 pathogen or by multiple pathogens in the setting. The distribution of the detected infections according to patients’ ages, genders, and seasons were also investigated.
MATERIALS AND METHODS
Study site and population
A cross-sectional study was undertaken from March 2016 through February 2017 in Taif region, western Saudi Arabia (Fig. 1). This city is located around 2,000 m above the sea level. The city has desert weather, with hot summers and mild winters. All months see some rainfall, with more rain reported in the spring and late fall months. Patients of different ages, those have been attending 3 primary health centers seeking treatment for their acute diarrheal episodes, were invited to participate in our study.
Inclusion/exclusion criteria
Acute diarrheal episode was defined as passage of ≥3 loose bowels over a day for a period ≤2 weeks before interviewing. Those patients who presented with diarrhea of ≥2 weeks duration and those received anti-parasitic or antibiotic medications for their diarrhea episodes, 2 weeks prior to the meeting, were excluded from the study. Following these criteria, 163 patients were recruited in the current study.
Ethical considerations
Patients or their relatives were informed of the aim of the study and signed consent forms authorizing their voluntary participation. The regional ethics committee approved data collection, clinical samples collection, and analysis of the study results.
Sample collection and processing
A total of 163 fresh stool samples, 1 sample per each patient, were collected in screw-capped appropriate containers and delivered within 2 hr to the microbiology laboratory at the College of Applied Medical Sciences, Taif University, Saudi Arabia. Upon arrival, each stool sample was inspected for consistency and for the presence of mucous or blood, carefully coded, and immediately stored at −20°C. On a monthly base, frozen samples were transported in ice packs to Al Borg central reference laboratory for molecular diagnosis with the xTAG™ GPP assay.
Molecular testing with xTAG GPP
From a single fecal specimen, the GPP assay allows identification of 15 potential enteropathogens (types, genotypes, and toxin-production genes) frequently-associated with diarrhea. The sought-for GI pathogens included Cryptosporidium species (C. parvum and C. hominis) Entamoeba histolytica, Giardia duodenalis, Campylobacter species (C. jejuni, C. coli, and C. lari), Clostridium difficile toxin A/B, Escherichia coli O157, enterotoxigenic Escherichia coli (ETEC) LT/ST strains, Salmonella., Shiga-like-toxin producing E. coli (STEC) stx1/stx2 strains, Shigella species (S. boydii, S. sonnei, S. flexneri, and S. dysenteriae), Vibrio cholerae, Yersinia enterocolitica, adenovirus subtypes 40/41, norovirus genogroups I and II (GI/GII), and group-A rotavirus.
Samples preparation and nucleic acid purification
Samples were initially prepared with the VERSANT Sample Preparation 1.0 Reagents kit (Siemens Healthcare Diagnostics, Erlangen, Germany) following the manufacturer’s protocol. The crude nucleic acids were directly retrieved from 700 μl of the fecal suspension using the automated platform VERSANT kPCR Molecular System. The purified nucleic acid samples were collected in 100 μl elusion buffer, following the manufacturer’s instructions.
Multiplex PCR reaction set up
The xTAG GPP assay was performed according to the manufacturer’s recommendations (Luminex Molecular Diagnostics, Austin, Texas, USA). Briefly, 10 μl of the nucleic acids extract together with the target-specific tagged primers and the biotin-labeled primers were initially used to set up the multiplex reverse transcription PCR (RT-PCR). The PCR reactions were carried out in Biometra T3 Thermocycler (Biometra GmbH, Göt-tingen, Germany). Amplification of the target DNA results in PCR amplicons with molecular weights ranging from 58 to 293 bp.
Bead hybridization reactions
Aliquots of the xTAG bead mix, 20 μl each, were subjected to sonication, applied into a 96-well microtiter plate, and 5 μl of each PCR product was added into specified well. Subsequently, the xTAG 0.22 streptavidin, R-phycoerythrin conjugate (SAPE), was diluted with the xTAG reporter buffer, and 75 μl of the solution was added into each well.
Products detection and analysis
Following hybridization, the median fluorescence intensity was generated for each xTAG bead population using the Luminex 100/200 instrument pre-heated to 45°C. The data were analyzed using the xTAG Data Analysis Software for the GPP assay.
Data analysis
The xTAG GPP assay results were tabulated for statistical analysis. The GI pathogen prevalence rate was calculated in Microsoft Excel and stated as a percentage of positive samples in relation to the total number of sample tested. The chi-square test was used to test associations between age groups, gender, and GI pathogens positivity variables. For<5 values, Fisher’s exact chi-square test was used, and the P-value<0.05 was considered significant.
RESULTS
Overall results
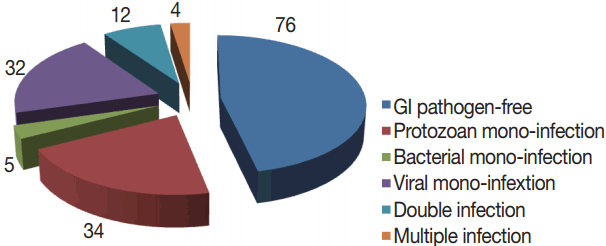
Total 163 patients were allocated in the current study; 91 were males and 72 were females. The age range was 0.5–60 years, with a mean age of 29.9±18.9 with the median age of 19. GI pathogens, 15 kinds, were looked-for in 1 stool sample collected from each participant with the xTAG GPP test. At least 1 enteropathogen was detected in 87 samples, with an estimated detection rate of 53.4%. Mono-infections were found in 71 (43.5%) samples and co-infections were identified in 16 (9.8%) samples. Moreover, co-infections included 12 (7.4%) samples presented as double infections and 4 (2.4%) samples were existed as triple infections (Fig. 2).
Spectrum of detected enteropathogens
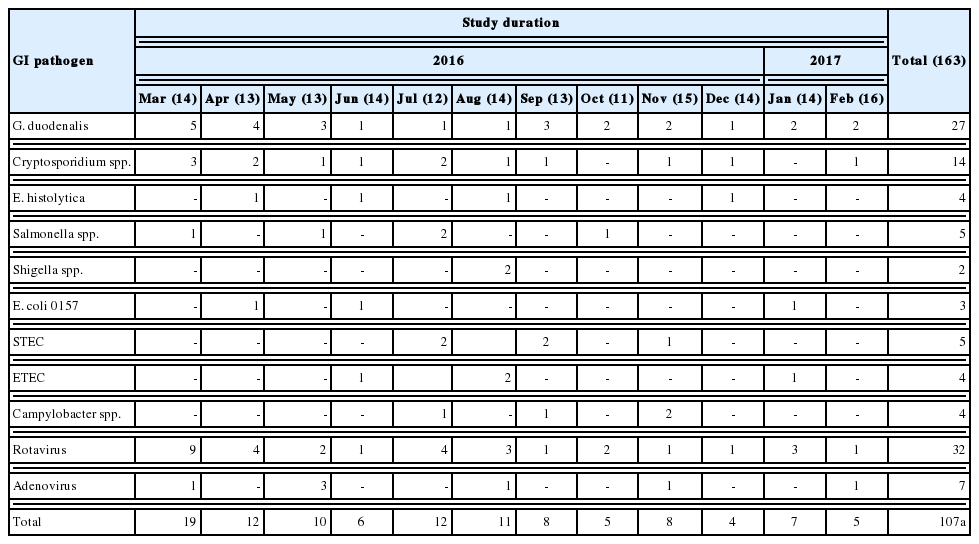
Among the 87 diarrheal episodes that were proved infectious, 34 (20.8%) were protozoan mono-infections, 32 (19.6%) were viral mono-infections, 5 (2.8%) were sole bacterial infections, and 16 (9.8%) were mixed infections (Fig. 2). Rotavirus (19.6%; n=32), G. duodenalis (16.5%; n=27), and Cryptosporidium spp., (8.5%; n=14), were the most commonly detected pathogens. Less frequently detected pathogens were adenovirus 40/41 (4.2%; n=7), Salmonella (3%; n=5) and STEC (3%; n=5), E. histolytica (2.5%; 4), ETEC (2.5%; 4), Campylobacter spp., (2.5%; 4), E. coli 0157 (1.8%; n=3), and Shigella (1.2%; n=2). No sample was positive for norovirus genotypes I and II (GI/GII), Vibrio cholerae, Yersinia enterocolitica, or Clostridium difficile toxin A/B (Table 1).
Pathogens involved in co-infections
All the detected pathogens were involved in co-infections except E. histolytica. Moreover, both G. duodenalis (5.5%; n=9) and rotavirus (3%; n=5) were the most predominant pathogens in co-infections. Campylobacter spp. (2.4%; n=4), ETEC (2.4%; n=4), E. coli 0157 (1.8%; 3), and Shigella spp., (1.2%; n=2) were detected in patients only as co-infections with other pathogens. Pathogens involved in co-infections were mentioned with their numbers in Table 1.
Distribution of pathogens relative to age and gender
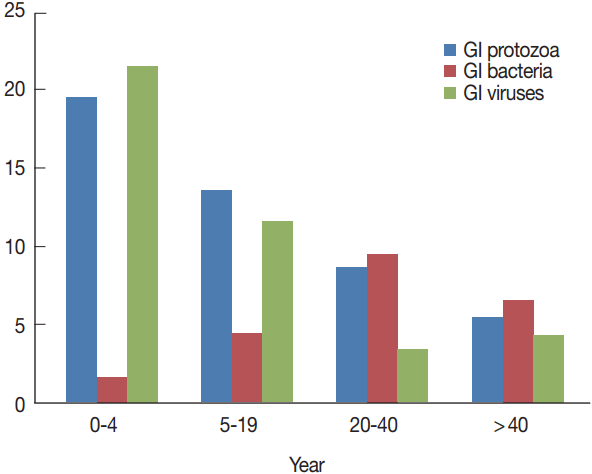
In general, infections were more in younger patients and in males, but no statistically significant difference observed between any of the 2 variables and the total number of pathogens detected (P =0.93). Children under 4 years (n=36) were the most infected patients, particularly for protozoa and viruses (Fig. 3). There were statistically significant differences among the distinct age groups and rotavirus infection (P < 0.001), Giardia infection (P =0.006), and Cryptosporidium infections (P =0.036). On the contrary, bacterial infections were seen more prevalent in patients aged 20–40 years and to a lesser extent in patients above 40 years, with no statistical significant differences stated among the different age groups and any identified bacteria (Table 1).
Month-wise distribution of detected pathogens
Table 2 shows the monthly distribution of the identified infection over 1 year period. Overall, infections occurred all the year around. Infections were dominant between March and August (70/107), including co-infections. GI protozoa were identified more in samples collected between March and June (22/45), the spring months in Saudi Arabia. Likewise, enteric viruses were detected more in samples collected during the same period (20/39), including co-infections. On the contrary, bacteria were identified more in samples collected in July and November (16/23), the summer months in the country. Lastly, none of the samples collected in December or October were positive for any enteric bacteria (Fig. 4).
DISCUSSION
To the best of our knowledge, this is the first study, in Saudi Arabia, concerned with detection of multiple GI pathogens, commonly recognized to cause diarrhea in humans using the xTAG™ GPP assay. Also, this is the first report in the Taif region that studies, in an all-inclusive manner, the common viral, protozoan, and the bacterial etiologies of diarrhea.
One noteworthy finding in our study was the exceptionally high number of enteric pathogens detected in this setting. Pathogens were detected in 87 out of 163 patients, with an estimated prevalence rate of 53.4%, a prevalence has not been previously described in this setting. In other settings, variable estimates have been reported in the country. Fareid et al. [15] identified enteric parasites in 50.4% and bacterial isolates in 88% of 160 schoolchildren presented with diarrhea based on conventional microscopy and stool cultures. Moreover, estimates of 34–46% have been reported for viral, bacterial, and parasitic causes of diarrhea in children less than 5 years [16–18]. However, the difference in populations, targeted and the methodologies adopted in these studies make the comparison problematic. With the xTAG GPP, Kahlau et al. [10] have reported detection of enteropathogens, mostly not requested by the physician, with a frequency rate of 45% in a hospitalized population [10]. Also, Albert et al. [11] have reported that the xTAG GPP detected twice as many pathogens as the conventional assays in a study done in Kuwait.
Overall, young children were a most vulnerable group to infections in this setting, going with tremendous of reports in the literature. Moreover, infections were more in males than females, in the absolute numbers, consistent with a previous study [19] and inconsistent with another [20]. Identification of groups of patients most vulnerable to infections guides the health authorities for better disease management, infection control, and resource allocation.
Also in our study, we clearly demonstrated a variety of enteropathogens detected and displayed their proportions in the etiology of diarrhea in the population studied. Viral gastroenteritis was a leading cause of diarrhea, in line with the literature. Rotavirus was detected in 32 out of 163 (19.6%) patients studied. Again, little has been recently published regarding rotavirus infection in our setting. None the less, estimates of 4–46% have been reported for other populations [21,22]. The adenovirus was identified with a frequency rate of 4.2%, lower than an estimate (1.4%) has been reported by Tayeb et al. [23] in other Saudi population [23]. Interestingly, no cases for norovirus genotypes I and II (GI/GII) were identified, perhaps being most frequently a nosocomial infection [24].
Coincided with several reports, viral gastroenteritis was predominant in children under 4 years, with a statistical significance observed (P <0.001) for rotavirus infections. Moreover, infections were more in female than in male patients, but without statistical significance, consistent with an Australian survey [20]. Although, viral infection was detected all the year around, the high positivity rate (59.3%) was displayed in the spring months, consistent with a Canadian report [25].
Regarding protozoa, all the 3 protozoan parasites targeted were detected in patients. Giardia was detected in 27 patients, with an estimated prevalence rate of 16.5%. In the same setting, a prevalence of 8.5–15% has been reported in 2 earlier studies [26,27]. Cryptosporidium infection was identified with an estimated prevalence of about 9%, minimally lower than an estimate, about 10%, described in an earlier study based on an in-house PCR [13]. In developing countries, detection of this opportunistic protozoan in diarrheal patients has gained great concern reporting much higher estimates [28]. Noteworthy was that both Giardia and Cryptosporidium have been widely recognized as common waterborne infections and found contaminating desalted water and ground water, 2 common drinking water resources in the country [29]. On the contrary to the above protozoa, E. histolytica was found only in 4 patients, with an estimated frequency of 2.4%. Frequencies of 6–18% have been displayed for E. histolytica in other Saudi populations based on microscopic diagnosis [30,31]. Like other enteric pathogens, much higher estimates have been described for this protozoon in developing countries [32]. Noteworthy, microscopic diagnosis of such unicellular parasite has been widely recognized insensitive and could not differentiate between E. histolytica and the 2 other morphologically similar species, namely E. dispar and E. moshkovskii.
Regardless the type of protozoa, infection were most frequent in children 0–4 years, less in adult<40 years, and least frequent in patients >40 years. There was significant difference stated in risk of across age groups with Giardia (P =0.006) and Cryptosporidium infections (P =0.036). Furthermore, Giardia infection was more in females, while Cryptosporidium was more in males, but these differences were not statistically significant. Although protozoa were identified in patients throughout the year, infections especially with Giardia and Cryptosporidium were detected in a greatest number (20/45) in spring months, coinciding with an earlier report [13].
Among the 9 bacteria studied, Salmonella, STEC, ETEC, and Campylobacter spp., E. coli 0157, and Shigella were isolated, with an estimated prevalence rate of 10%. Estimates between 10% and 30% have been reported based on culture methods [16,18]. No cases for Vibrio cholerae, Yersinia enterocolitica, or Clostridium difficile toxin A/B were identified. Perhaps, measures taken by the health-authorities and increasing the awareness of the population about the importance of good food hygiene, after a period of foodborne diarrhea outbreaks, may be an explanation [33]. C. difficile was like norovirus commonly acquired in hospitals [34]. In contrast to the viral and protozoan infections, the enteric bacteria were identified more in teenagers and in males, with no statistical significance observed in risk across patients’ ages or genders with any of the bacterial isolates. Furthermore, more bacteria were detected in the summer months.
Also in this context, we clearly demonstrated the frequency of infections with multiple pathogens in patients with diarrhea. Co-infections were detected in 16 patients, with an estimated prevalence rate of 9.8%, comparable to estimates, 7.0–9.5%, documented, elsewhere [7,8]. Identical to several mono-infections detected, co-infection was identified more in children than other age groups and in males than females, but no significant statistical difference observed across both variables and the incidence of getting infected with multiple pathogens. Interestingly, all detected pathogens were found involved in mixed infections with except E. histolytica. E. histolytica has been frequently co-infected with the other 2 morphologically similar species E. dispar and E. moshkovskii, not covered by the xTAG GPP assay [35]. Both G. duodenalis and rotavirus were the 2 pathogens most frequently detected in mixed infections.
Interestingly, Campylobacter spp., ETEC, E. coli 0517, and Shigella spp. were identified in the studied population only as concomitant pathogens. Co-infections among enteric pathogens have been frequently described [36–38]. However, still there is a controversy regarding the clinical impact of concomitant infections on the diarrheal illness. Synergic effects of concomitant infections have been ruled out for rotavirus infection in a study and reported later in another [39,40].
Lastly, our study could be bounded by a number of factors. Some interesting data could be statistically biased by the relatively small number of positive samples. Moreover, non-inclusion of a control group in conjunction with patients did not help to clear the clinical impact of some findings. Lastly, pathogens commonly reported to cause diarrhea in the country like astrovirus, intestinal helminths, and other coccidian parasites, are not covered by the xTAG GPP assay and therefore, these enteric pathogens were not investigated in this study.
To conclude, in this report, we clearly demonstrated the high burden of pathogens causing diarrhea in the study setting. Giardia, rotavirus, and Cryptosporidium were the most dominant pathogens. Infections were frequent in children below 4 years. Furthermore, co-infections were identified with a relatively high frequency. Lastly, the methodology adopted in this study was proved useful for detection of multiple potential pathogens in this population. Further longitudinal case-control studies are needed to follow-up patients and define accurately the clinical impact of the identified pathogens on the diarrheal disease.
ACKNOWLEDGMENT
This research was funded by Taif University, Saudi Arabia (grant no. 1-437-4949). The authors would like to thank Dr. Rizk A, Dr. Henawy I, and their colleagues for their great help while recruiting patients and collecting fecal samples.
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this study.