Prevalence of Tick-Borne Pathogens from Ticks Collected from Cattle and Wild Animals in Tanzania in 2012
Article information
Abstract
This study was aimed to disclose the prevalence rate of tick-borne pathogens from ticks collected from cattle and wild animals in Tanzania in 2012. Ticks were collected from slaughtered cattle and dead wild animals from November 5 to December 23, 2012 and identified. PCR for detecting Anaplasmataceae, Piroplamidae, Rickettsiaceae, Borrelia spp., and Coxiella spp. were done. Among those tested, Rickettsiaceae, Piroplasmidae, and Anaplasmataceae, were detected in ticks from the 2 regions. Rickettsiaceae represented the major tick-borne pathogens of the 2 regions. Ticks from animals in Maswa were associated with a higher pathogen detection rate compared to that in ticks from Iringa. In addition, a higher pathogen detection rate was observed in ticks infesting cattle than in ticks infesting wild animals. All examined ticks of the genus Amblyomma were infected with diverse pathogens. Ticks of the genera Rhipicephalus and Hyalomma were infected with 1 or 2 pathogens. Collectively, this study provides important information regarding differences in pathogen status among various regions, hosts, and tick species in Tanzania. Results in this study will affect the programs to prevent tick-borne diseases (TBD) of humans and livestock in Tanzania.
Shinyanga Province, located in northern Tanzania, is bordered by Lake Victoria. Shinyanga Province has the largest livestock numbers in Tanzania [1]. Maswa Game Reserve of Shinyanga Province is located near Serengeti National Park, where-in cattle herds share habitats and grassland with wild animals. Iringa Province, near Ruaha National Park and Udzungwa National Park, is located in the highlands of south central Tanzania. The meat processing industry is developed in Iringa urban Province. High rates of tick infestation and TBD-associated mortality remain a major problem in these areas [2]. As livestock and wild animals share habitats and grassland, tick infestation is influenced by the transmission of various tick-borne pathogens among these animals.
We previously reported the identification of ticks in cattle and wild animals from Maswa and Iringa [1]. Amblyomma variegatum and A. gemma were determined to be the most common ixodid tick species associated with animals of Maswa and Iringa, respectively. In this study, we identified the TBD-causing pathogens transmitted from ticks that were collected from cattle and wild animals from these 2 areas. Results in this study can provide important information for the establishment of TBD prevention programs in these areas.
Ticks collected from the cattle and wild animals (buffalo, bush pig, eland, leopard, warthog, and zebra) from Maswa Game Reserve and Iringa urban, Tanzania from November 5 to 23, 2012 [1] were used in this study. Collected ticks were fixed with EtOH, and transferred from Tanzania to Yonsei University, Korea. They were frozen with liquid nitrogen and homogenized. Total DNA was isolated using a genomic DNA extraction kit (GeneAll, Seoul, Korea) according to the manufacturer’s instructions, and stored at −20°C until use. To detect Anaplasmataceae, Piroplasmidae, Rickettsiaceae, Borrelia spp., and Coxiella spp. in ticks from animals, PCR amplifications (from about 400 to 600 bp) targeting 16S rRNA, 18S rRNA, 17 kDa protein, flab, and htpB, respectively, were performed using primers and thermal cycling conditions described in a previous report [3]. Appearances and the sizes of PCR amplicons were the main criteria for the detection of the pathogens. Some PCR amplicons were directly purified using a PCR purification kit (GeneAll), and sequenced (Genotech, Daejeon, Korea). Sequences were compared to those available in GenBank by performing a NCBI BLAST search. The data obtained in this study were analyzed using SPSS version 23.0 (SPSS, Chicago, Illinois, USA). Differences in pathogen detection were tested by a Fisher’s exact test. Significance (for all results) was considered at P ≤0.05.
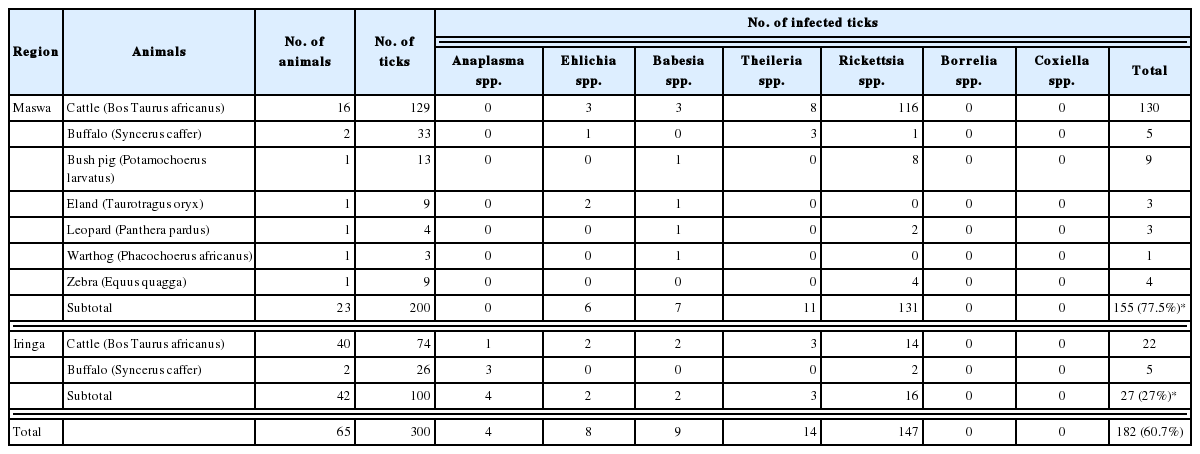
Among the 300 ticks examined, pathogens were detected by PCR in 187 (60.7%) ticks (Table 1). Ticks from animals of Maswa exhibited a 77.5% pathogen detection rate, which was higher than that for ticks from animals from Iringa (27%). Among members of the Anaplasmataceae family, uncertain Anaplasma spp. were detected in only 4 ticks, which were from cattle of Iringa. Ehrlichia spp. (E. ruminatium and E. chaffeensis) were detected in ticks from cattle, buffalo, and eland from both regions. In total, 4% (12/300) of ticks were infected with Anaplasmataceae in animals from both regions. Nine ticks from cattle, bush pig, eland, leopard, and warthog from Maswa were infected with Babesia spp. (B. caballi and uncertain Babesia sp.). In addition, Theileria spp. (T. velifera and T. mutans) and uncertain Theileria sp. were detected in 14 ticks from cattle and buffalo of both regions. Rickettsiaceae was the most common family of pathogens identified in ticks from both regions. Rickettsiaceae (R. africae, R. rickettsii, and uncertain Rickettsiaceae) was detected in 146 ticks, and from all animals examined except warthog. Rickettsiaceae detection rates were 65.5% (131/200) and 16% (16/100) in ticks from animals of Maswa and Iringa, respectively. Cattle was the most common host animal of ticks harboring Rickettsiaceae. Finally, Borrelia and Coxiella spp. were not detected in this study.
Mixed infections were detected in 17 ticks (5.67%) (Table 2). Double infections by Rickettsiaceae and Piroplasmidae were the most common type of mixed infection. Triple infections (Babesia spp., Theileria spp., and Rickettsiaceae) were found in 2 ticks from Maswa cattle. Only 2 ticks from wild animals of Maswa had mixed infections. Generally, the pathogen detection rate in ticks from cattle was 2-fold higher than that in ticks from wild animals.
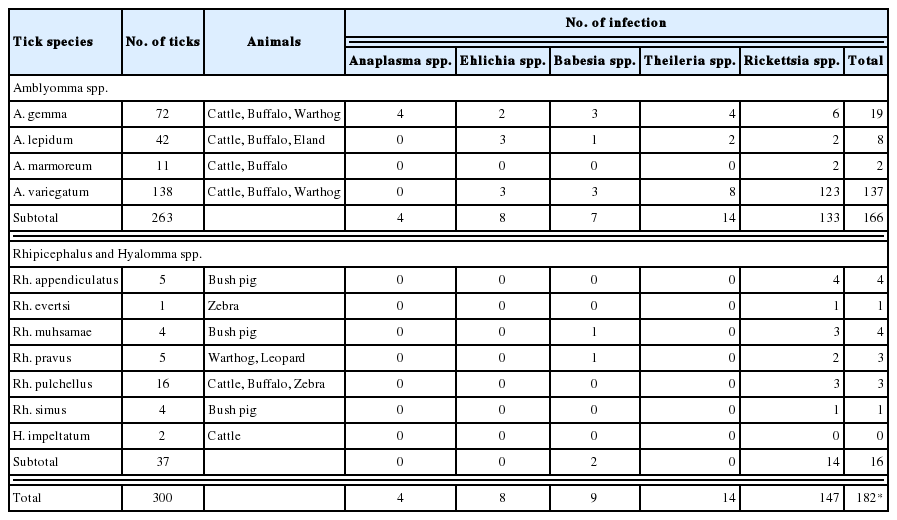
In general, Amblyomma ticks were associated with a higher pathogen detection rate than Rhipicephalus and Hyalomma ticks (Table 3). Due to the high Rickettsiaceae detection rate (123/137), almost 100% A. variegatum ticks [1], the most predominant tick species, were infected with pathogens. The second most predominant species, namely A. gemma, was associated with a 25% (18/72) pathogen detection rate. Amblyomma ticks were infected with diverse pathogens, whereas Rhipicephalus and Hyalomma ticks were generally infected by a single Rickettsia species. Exceptionally, Rhipicephalus muhsamae and Rhipicephalus pravus ticks were infected with Babesia spp. as well as Rickettsia spp.
The pathogen detection rate in ticks collected from animals of the Maswa region was higher than that in ticks from the Iringa region. In general, the pathogen detection rate in ticks from cattle was 2-fold higher than that in ticks from wild animals. This is due to the high pathogen detection rate in A. variegatum, which was the predominant tick species associated with animals, especially cattle, in Maswa region. The detection rate of pathogens in A. variegatum was much higher than that in A. gemma, which was predominant in Iringa. Being dominant in cattle, the migration of A. variegatum, which transmits pathogens among cattle, in nearby areas is a common event in the herds of Maswa. Among the pathogens identified in this study, Rickettsiaceae was the most common in ticks from both regions. A high Rickettsiaceae detection rate in A. variegatum from the cattle of Maswa contributed to a 3-fold higher Rickettsiaceae detection rate in the animals of Maswa (Table 1). The detection rate of pathogens other than Rickettsiaceae in ticks from Maswa (12.0%, 24/200) and Iringa (11.0%, 11/100) were similar. Only ticks from cattle of Iringa were associated with Anaplasma sp. infection. Generally, higher pathogenic diversity in ticks from cattle was observed in Iringa.
In a previous report, more diverse ticks of genus Rhipicephalus were collected from wild animals [1]. Pathogens other than Rickettsiaceae and Babesia spp. were not shown to infect Rhipicephalus ticks in this study. Since the habitats of wild animals are larger than those of cattle, Rhipicephalus ticks might be considered good environmental factors for the widespread distribution of Rickettsiaceae infection.
In this study, the detection rates of Anaplasmataceae and Piroplasmidae were very low compared to those of Rickettsiaceae. However, the detection rates of Anaplasma marginale and Babesia bigemina in bovine blood in Iringa reached 37% and 43%, respectively [4,5]. Therefore, more extensive studies on tick-mediated Anaplasmataceae and Piroplasmidae infection are required.
Unlike Rickettsiaceae, Anaplasmataceae, Piroplasmidae and other pathogens, Borrelia and Coxiella spp. were not detected in ticks in this study. However, the possibility of diseases caused by these genera remains high in Tanzania, based on the detection of antibodies against Borrelia and Coxiella spp. in cattle and human sera [6–9]. The possibility that Arthropods other than ticks can transmit Borrelia and Coxiella spp. in Tanzania should also be considered [10–12].
This study is the first report of ticks and tick-borne pathogens in cattle and wild animals of Maswa and Iringa regions, Tanzania. We focused on comparing pathogens between the 2 regions, between cattle and wild animals, and among tick species. Since livestock and wild animals share habitats and grassland in these regions, studies are necessary to determine if ticks can transmit pathogens between livestock and wildlife, and even to humans in the region. This study provides very important information and basic knowledge that is necessary to initiate TBD prevention programs in these areas.
ACKNOWLEDGMENT
This study was supported by the Project of Cooperation on Bioresources with Tanzania Wildlife Research Institute (TAWIRI) through the National Research Foundation (NRF) of Korea (2012K1A3A9A01027507).
Notes
CONFLICT OF INTEREST
All authors declare that they have no conflict of interest to this study.