Biological Characteristics of Recombinant Arthrobotrys oligospora Chitinase AO-801
Article information
Abstract
Chitinase AO-801 is a hydrolase secreted by Arthrobotrys oligospora during nematode feeding, while its role remained elusive. This study analyzed the molecular characteristics of recombinant chitinase of Arthrobotrys oligospora (reAO-801). AO-801 belongs to the typical glycoside hydrolase 18 family with conserved chitinase sequence and tertiary structure of (α/β)8 triose-phosphate isomerase (TIM) barrel. The molecular weight of reAO-801 was 42 kDa. reAO-801 effectively degraded colloidal and powdered chitin, egg lysate, and stage I larval lysate of Caenorhabditis elegans. The activity of reAO-801 reached its peak at 40°C and pH values between 4–7. Enzyme activity was inhibited by Zn2+, Ca2+, and Fe3+, whereas Mg2+ and K+ potentiated its activity. In addition, urea, sodium dodecyl sulfate, and 2-mercaptoethanol significantly inhibited enzyme activity. reAO-801 showed complete nematicidal activity against C. elegans stage I larvae. reAO-801 broke down the C. elegans egg shells, causing them to die or die prematurely by hatching the eggs. It also invoked degradation of Haemonchus contortus eggs, resulting in apparent changes in the morphological structure. This study demonstrated the cytotoxic effect of reAO-801, which laid the foundation for further dissecting the mechanism of nematode infestation by A. oligospora.
INTRODUCTION
Gastrointestinal nematode diseases in sheep are common and highly prevalent parasitic diseases caused by various gastrointestinal nematodes (GINs). Infection with GINs is characterized by emaciation, diarrhea, anemia, poor mental health, and even death in sheep [1,2]. Currently, the treatment options available for gastrointestinal nematode disease in sheep primarily include chemical prevention and chemotherapeutic control. However, the limitations of chemical anthelmintics, such as nematode resistance, environmental pollution, and food safety, are becoming a major concern in the prevention of the disease [2,3]. Therefore, biological control using nematophagous fungi has gained the focus of researchers for the prevention and control of gastrointestinal nematode diseases in sheep.
Arthrobotrys oligospora is a nematophagous fungus widely found in the natural habitat. It is currently the most promising fungus used for biological control [1,4]. In response to the environmental stress or nematode stimulation, A. oligospora produces a specific predatory structure, precisely, a sticky web to capture the nematodes. It also secretes extracellular hydrolases and participates in nematode immobilization, cuticle invasion, and nematode cell degradation [4–8]. A. oligospora-secreted extracellular hydrolases mainly include serine proteases and chitinases. The serine proteases PII, VCP1, Aoz1, P186, and Ver112 are known to degrade the nematode cuticle [6,9,10]. In addition, the secreted chitinases are involved in degrading the chitinous component of the nematode eggs and the cell wall of pathogenic fungi [5,8,11–13]. Generally, the nematode eggshell is made up of the lipid layer, chitinous layer, and vitelline layer, of which chitin accounts for approximately 40 %, and is the thickest layer of the eggshell that constitutes the primary barrier to infection [14]. A chitinase Chi43 from Pochonia chlamydosporia showed that a degradation effect on Globodera pallida eggs [15]. The application of Paecilomyces lilacinus chitinase to Meloidogyne javanica eggs resulted in significant changes in the eggshell structure and fragmentation of the yolk layer, resulting in loss of integrity [16]. The chitinase of Lecanicillium antillanum accelerates the softening process of Meloidogyne incongnita eggs, degrade the eggshell, and reduce the permeability of the eggshell, thereby interfering with the hatching of nematode eggs or making the larvae break early [17]. The nematophagous fungi chitinases can prevent and control nematode infections by remarkably reducing hatching rate of nematode eggs. The chitinases can serve as an indicator for screening high-efficiency nematophagous fungi strains [8,11,12].
Few studies are available on the chitinase of A. oligospora. Previous studies have reported that the chitinase of A. oligospora plays a crucial role in nematode infection [8,11,12]. Yang et al. [6,8] sequenced the whole genome of A. oligospora and subsequently performed transcriptional analysis under different culture conditions. The gene of AOL_s00215g801 (AO-801) transcription was significantly up-regulated under nitrogen starvation conditions or in the presence of chitin, indicating that the chitinase AO-801 might play a crucial role in nematode infestation by A. oligospora [8]. However, a comprehensive investigation of the structure and function of AO-801 has not been addressed. This study explored the molecular characteristics, enzymatic characteristics, and nematicidal activity of AO-801. Our findings provide new insights into the role of AO-801 and mechanisms underlying the infestation of nematodes by A. oligospora via AO-801.
MATERIALS AND METHODS
Ethical approval
All experiments were carried out in accordance with the guidelines issued by the Ethical Committee of Shihezi University (No. A20180126).
Fungi strains
A. oligospora XJ-A1 strain was isolated and preserved by the Laboratory of Parasitology, College of Animal Science and Technology, Shihezi University (Xinjiang, China). The vectors, pMD19-T and pPIC9K were purchased from Takara and Invitrogen, respectively. Escherichia coli DH5α and Pichia pastoris GS115 were purchased from Invitrogen.
Primers
AO-801 gene-specific primers P1 (5′-GGAATTCATGAAGGCCATCTATGGACGTAACT-3′) and P2 (5′-CGAGCGGCCGCTCAAGCGCAAGCGCTGCG-3′) were designed by software Premier 5.0 based on the GenBank-published sequence AOL_s00215g801 of the standard strain of A. oligospora (ATCC 24927) (underlined sequences are the restriction sites for EcoR I and Not I, respectively). The primers were synthesized by BGI Sequencing (Beijing, China).
Total RNA extraction and cDNA synthesis
Conidia of A. oligospora XJ-A1 were inoculated on the potato dextrose medium (PDB) and incubated at 28°C on a constant shaker at 80 rpm (protected from light) to enrich the mycelia. After 48 h incubation, stage-III larvae of Caenorhabditis elegans were transferred onto the mycelia. Subsequently, the mycelia were harvested by filtering through the sterile filter paper, after the appearance of a large number of sticky webs. Finally, total RNA was extracted using EZ-10 Spin Column Fungal RNA Mini-Preps Kit (Bio Basic Inc, Ontario, Canada), following the manufacturer’s instructions. Reverse-transcription to cDNA was performed using the PrimeScript RT kit (Takara, Tokyo, Japan).
Cloning and molecular characterization
The cDNA was used as a template to amplify AO-801 gene. PCR amplification products were isolated and recovered using an agarose gel DNA extraction kit (Takara). Then, the amplified products were cloned into the pMD19-T vector to construct the recombinant plasmid pMD19-AO801. Further, the recombinant construct was transformed into E.coli DH5α competent cells to screen positive clones for sequencing. The signal peptide was predicted from the amino acid sequence using the online tool, Signal P (http://www.cbs.dtu.dk/services/Signal P/). The protein hydrophilicity was analyzed using ProtScale (http://expasy.org/tools/protscale.html). In addition, SWISS-MODEL (https://www.swissmodel.expasy.org/) was used to predict the tertiary structure of the chitinase. Further, sequence alignment of the chitinases was performed using DNAMAN. Finally, phylogenetic analysis was performed using MEGA 6.0, a neighbor-joining (NJ) tree was drawn, and bootstrap analysis was set to 1,000 replicates [8].
Expression, identification, and purification
The plasmids pMD19-AO801 and pPIC9K were digested with the restriction enzymes EcoR I and Not I (Takara), respectively. Next, the linearized pPIC9K vector and the digested AO-801 gene fragment were ligated to construct the recombinant expression vector pPIC9K-AO801. After verification using the restriction digestion with EcoR I and Not I, pPIC9K-AO801 was linearized with Stu I (Takara) and transformed into the Pichia pastoris GS115 competent cell by electroporation. The transformed yeast cells were screened using geneticin (Takara). Subsequently, the transformed yeast cells were inoculated on a BMGY medium, and after culturing for 24 h at 28°C, the cells were harvested by centrifugation at 2,500 rpm for 10 min. These cells were resuspended in a BMGY medium, and methanol was added every 24 h to obtain a final concentration of 1%. The supernatant obtained after 24, 48, 72, and 96 h of induction were subjected to SDS-PAGE, and stained using Coomassie brilliant blue staining. Western blot analysis was performed using mouse anti-AO-801 (prokaryotic expression) positive serum as a primary antibody (dilution ratio 1:1,000) and HRP-goat anti-mouse IgG (TransGen, Beijing, China) as the secondary antibody (dilution ratio 1:2,000). Finally, purification of recombinant chitinase AO-801 (reAO-801) was performed using a Ni-NTA purification system (Solarbio, Beijing, China).
Enzymatic activity
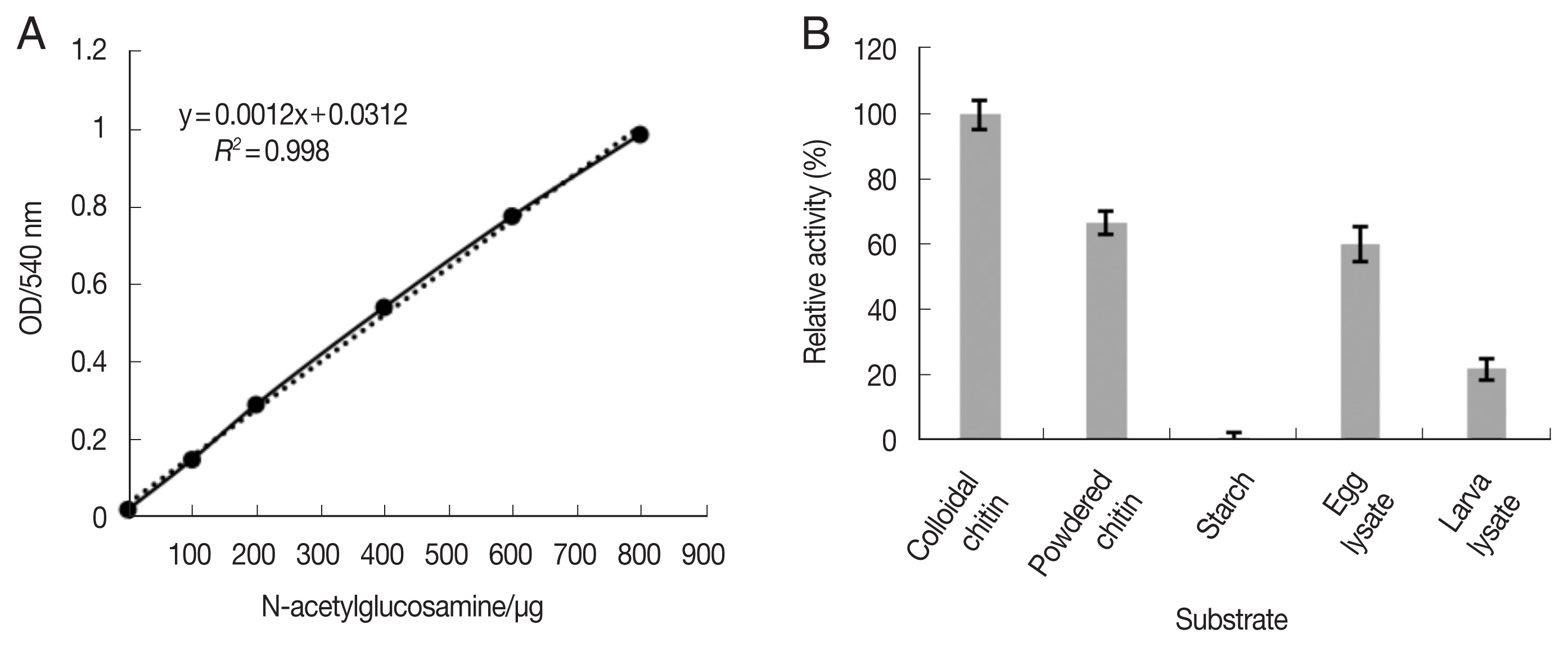
The enzymatic activity of reAO-801 was analyzed by following the method described by Berger et al. [18] for the measurement of reducing sugars. In addition, the absorbance values of reaction products were measured at OD540 nm using a full-wavelength microplate reader (Thermo Scientific, Madison, Wisconsin, USA). The standard curves were plotted in Microsoft Excel. One unit of enzyme activity was defined as the amount of enzyme that produces 1 mg of reducing sugar in 1 min.
Substrate specificity
About 100 μl of reAO-801 was mixed with different substrates, such as 400 μl of colloidal chitin solution (pH 7.0), powdered chitin (RYON, Shanghai, China) suspension, starch (Oxoid, Basingstoke, England) solution, C. elegans egg lysate and stage-I larvae lysate of C. elegans. The mixed solution was incubated at 37°C for 1 h, after which 500 μl of 3,5-dinitrosalicylic acid (DNS) (Biopeony, Beijing, China) reagent was added. The absorbance value of the supernatant (200 μl) was measured at 540 nm.
Effects of temperature, pH, metal ions, and chemical substances
The effects of different temperatures, pH, metal ions, and chemical substances on the enzymatic activity of reAO-801 were determined separately, as described in the previous section. Briefly, reAO-801 and colloidal chitin solution (pH 7.0) were mixed and incubated at different temperatures to determine the relative activity. Next, reAO-801 was mixed with colloidal chitin solutions of different pH values to determine the effect of pH on the relative activity. Further, 10 μl of metal ion solution (1 mol/L), such as zinc sulfate solution, magnesium chloride solution, calcium chloride solution, iron chloride solution, and potassium chloride solution, was mixed separately with reAO-801 and kept at 4°C for 1 h. Then, the mixed solution was incubated with colloidal chitin solution to determine the relative activity. In addition, 25 μl of urea (0.5 mol/L), ethylenediaminetetraacetic acid (EDTA) (0.05 mol/L), 1% sodium dodecyl sulfate (SDS), and 2-mercaptoethanol (0.05 mol/L) were mixed separately with reAO-801 and kept in room temperature for 5 min and then incubated with colloidal chitin solution (pH 7.0) to determine the relative activity.
Degrading activity of reAO-801 against nematodes and their eggs
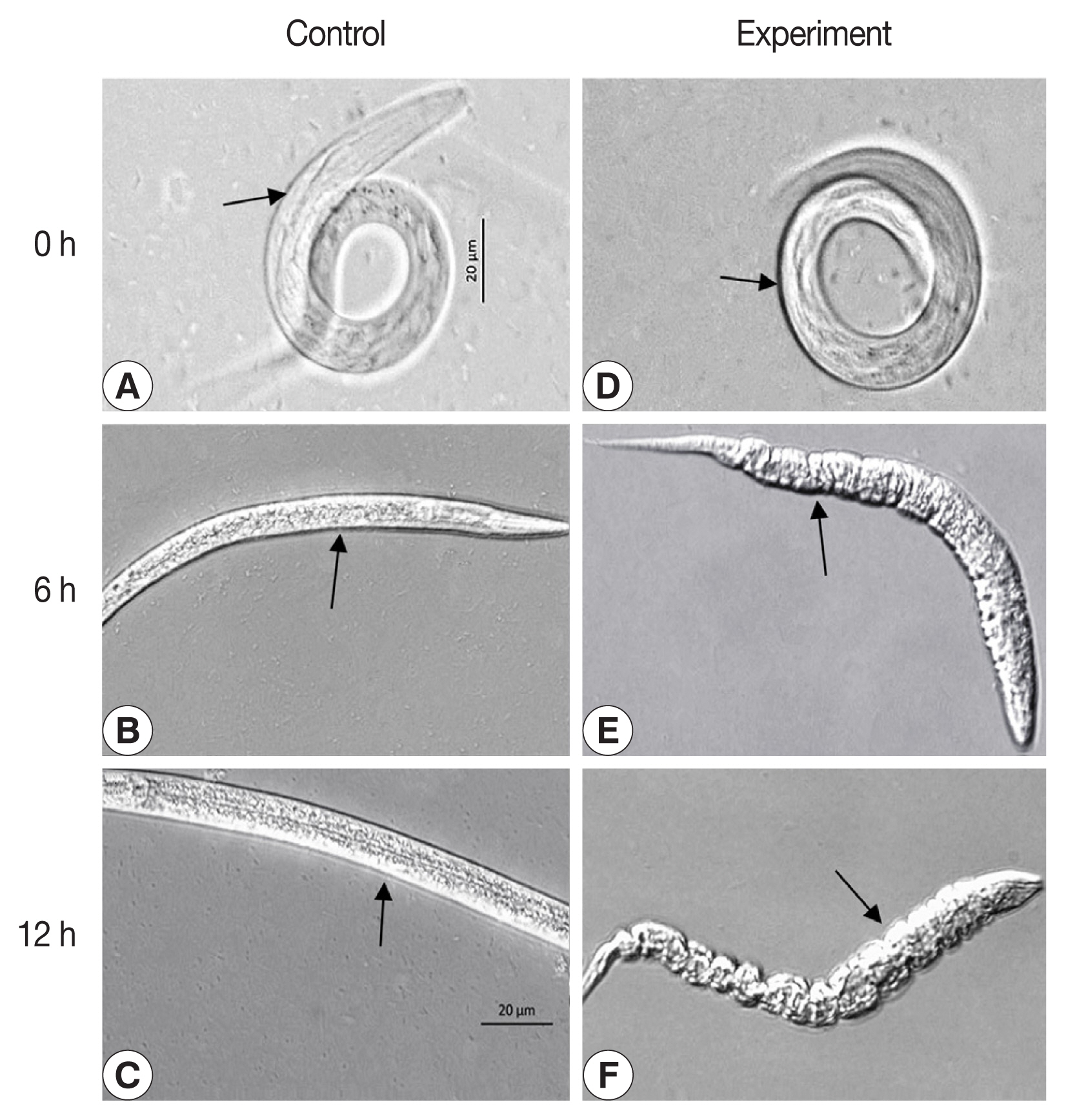
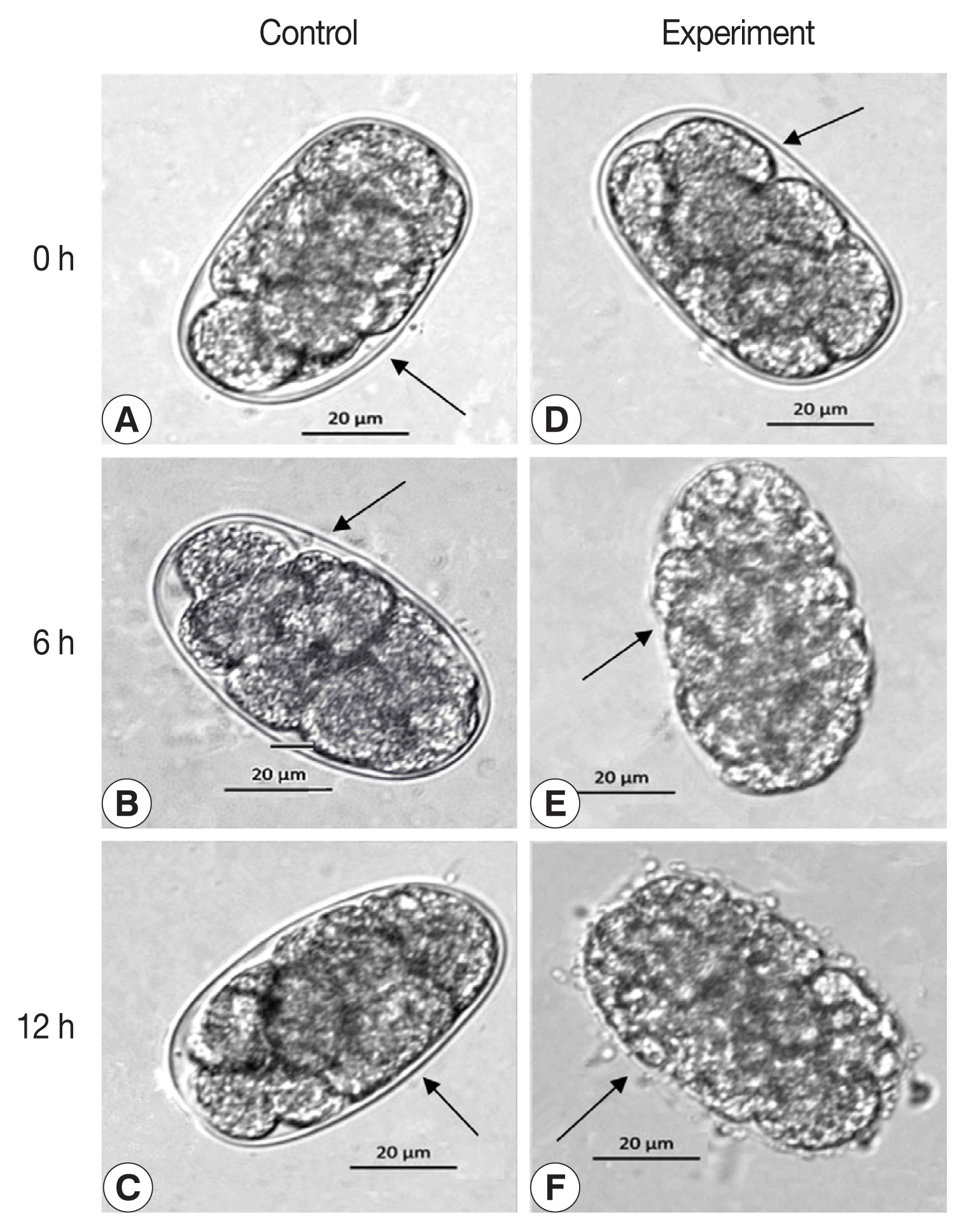
Initially, 400 μl of reAO-801 was mixed separately with approximately 1,000 nematodes or their eggs, such as C. elegans larvae, C. elegans eggs, and Haemonchus contortus (isolated from naturally infected abomasum of sheep) eggs, and incubated at 37°C for 0, 6, and 12 h. Subsequently, morphological parameters were observed with an inverted microscope (ZEISS AxioObserverA1, Jena, Germany). The high temperature-inactivated AO-801 (incubated at 100°C water bath for 10 min) was used as a corresponding control to analyze the degradation activity of reAO-801 against nematode larvae and their eggs.
Statistical analysis
The absorbance value of heat-inactivated AO-801 in the reaction mixture was used as blank controls. The enzymatic activity of the reaction mixture containing reAO-801 and colloidal chitin (pH 7.0) at 37°C was defined as 100%. All the experiments were repeated 3 times. Data were analyzed and plotted in the form of a graph using Microsoft Excel. Finally, an analysis of variance (ANOVA) was performed to evaluate the statistical significance.
RESULTS
Cloning and molecular characterization of AO-801 gene
The open reading frame of the AO-801 cDNA was 1,131 bp in length and encoded 376 amino acids (Supplementary Fig. S1). No signal peptide was predicted. It showed 97.34% homology with the amino acid sequence of AO-801 of A. oligospora ATCC 24927. AO-801, which belongs to the glycoside hydrolase 18 family, had a Glyco-18 domain (amino acids 3 to 333) and 2 highly conserved regions, SIGGW and LDGVDVDWE. SIGGW was located at the 82nd-86th amino acid residues, which were the substrate-binding sites, and hydrolase catalytic active site constitutedLDGVDVDWE was located at the 120th-128th amino acid residues (Supplementary Fig. S2A). SWISS-MODEL suggested that the tertiary structure of AO-801 had a triosephosphate isomerase TIM barrel of (α/β)8 (Supplementary Fig. S2B). Homology analysis revealed that AO-801 exhibited the highest homology (71.43%) with the chitinase of Dactylella cylindrospora (KAF3934333) (Supplementary Fig. S3) and shared the evolutionary branch with the chitinase of plant pathogenic fungi (Supplementary Fig. S4).
Expression, purification, and identification of reAO-801 protein
The recombinant vector pPIC9K-AO801 was constructed for the expression of reAO-801 in yeast cells (Supplementary Fig. S5). We obtained recombinant yeast cells resistant to G418 (Supplementary Fig. S6). When the recombinant yeast was induced by 1% methanol for 120 h, the recombinant protein concentration reached the maximum in the extract (0.99 mg/ml). SDS-PAGE analysis revealed that the expressed protein was consistent with the expected protein size, with a molecular mass of about 42 kDa (Supplementary Fig. S7A). Western blot analysis showed that specific binding with the mouse anti-AO-801 anti-serum (Supplementary Fig. S7B). We obtained 4.52 mg/ml of the purified protein.
Different substrates for reAO-801
The N-acetylglucosamine (NAG) standard curve based on the method used for reducing sugar measurement met the experimental requirements (Fig. 1A). reAO-801 was found to be substrate-specific, but showed some differences in the degradation activity across different substrates. The degradation activity of reAO-801 to colloidal chitin was the strongest, and the activity to powdered chitin, C. elegans egg lysates and stage-I larval lysates containing chitin were successively weakened. Altogether, reAO-801 could effectively degrade colloidal chitin, powdered chitin, egg lysates, and stage-I larval lysates of C. elegans, but not starch (Fig. 1B).
Effects of temperature, pH, metal ions, and chemical substances on the enzymatic activity of reAO-801
The enzymatic activity of reAO-801 was high at 30–50°C and highest at 40°C, but at 70°C, it was almost completely inactivated (Fig. 2A). reAO-801 enzymatic activity was high at pH 4–7 and peaked at pH 6 (Fig. 2B). Zn2+, Ca2+, Fe3+ inhibited its activity, while Mg2+, K+ promoted its activity; however, the effects were not significant (Fig. 2C). Urea, SDS, and 2-mercaptoethanol could significantly inhibit the activity of reAO-801, decreasing its activity to less than 5% compared to the control. On the contrary, the inhibitory effect of EDTA on the reAO-801 was minimal (Fig. 2D).
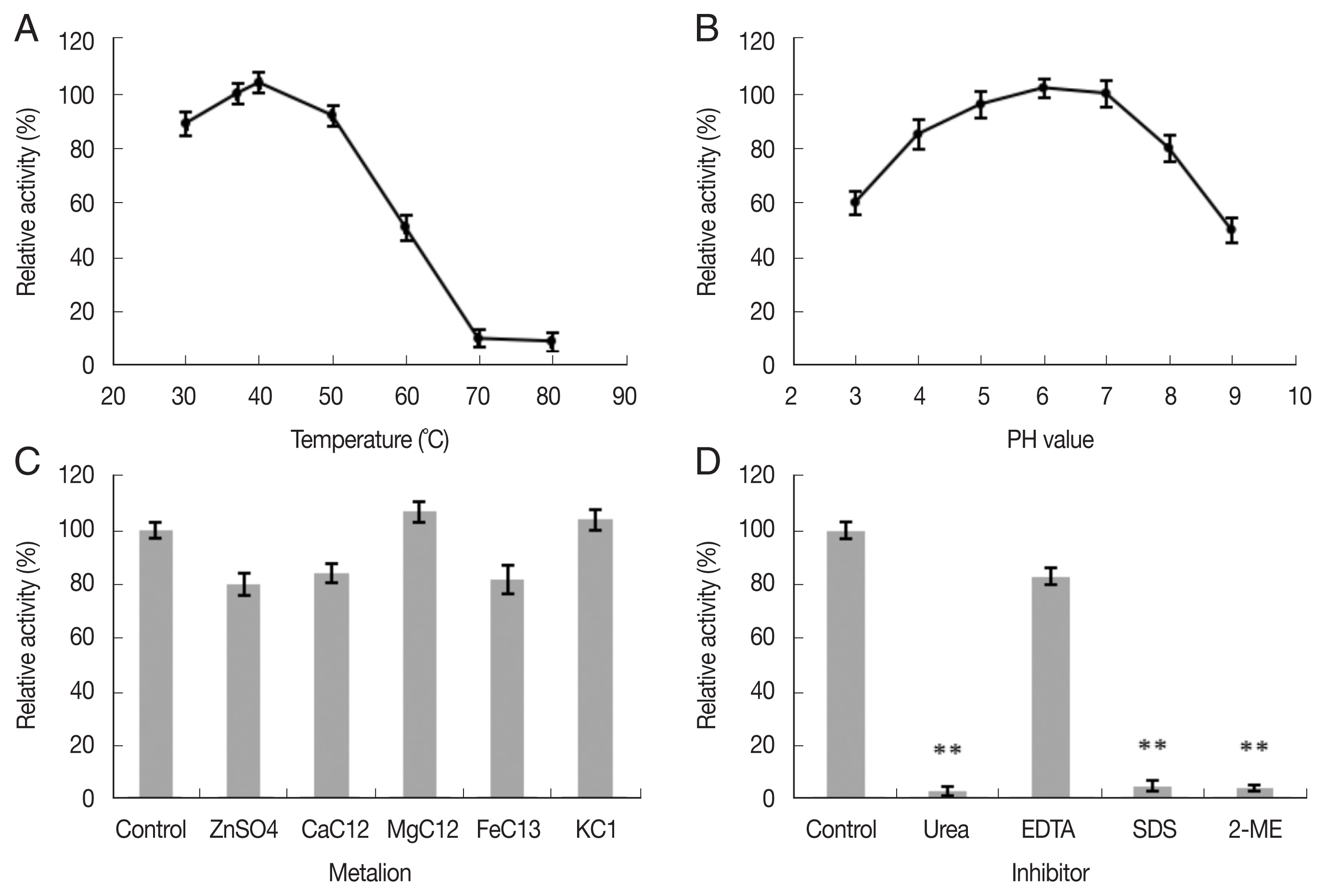
Effects of temperature, pH, metal ions and inhibitors on reAO-801. (A) Effects of different temperature on reAO-801. (B) Effects of different pH on reAO-801. (C) Effects of different metal ions on reAO-801. (D) Effects of different inhibitors on reAO-801.Relative activity was expressed as a percentage of the standard activity. All values were the average of 3 replications. Error bars represents the standard deviations. **P< 0.001.
Degradation activity of reAO-801 against stage-I larvae of C. elegans
Compared to the control group (Fig. 3), the larval viability of C. elegans treated with reAO-801 decreased; the mortality rate reached 62% and 100% after 6 h and respectively 12 h. The body wall of dead nematodes in the experimental group contracted and lost elasticity.
Degradation activity of reAO-801 against the eggs of C. elegans and H. contortus
The egg development in C. elegans was halted at 6 h following exposure to reAO-801 compared to that in the control group (Fig. 4). The larvae inside the eggs of the experimental group did not circle or twist, the wall of the eggshell became siginificantly thinned, and the structure of the eggshell collapsed or even disappeared (Fig. 4E). After 12 h exposure, the eggshell completely disappeared. The larvae inside the eggs died and exhibited irregular shapes (Fig. 4F). The shells of some eggs of the experimental group broke down, leading to the premature release of the larvae with lower activity (Fig. 4H). The larvae hatched early in the experimental group died completely 12 h following exposure, and the body walls were crumpled (Fig. 4I). In addition, the eggs of H. contortus of the experimental group ceased to develop 6 h following reAO-801 treatment and the eggshells disappeared significantly, resulting in non-clear eggshell structure boundaries (Fig. 5). By 12 h, the eggshells had disappeared and the egg contents were completely exposed.
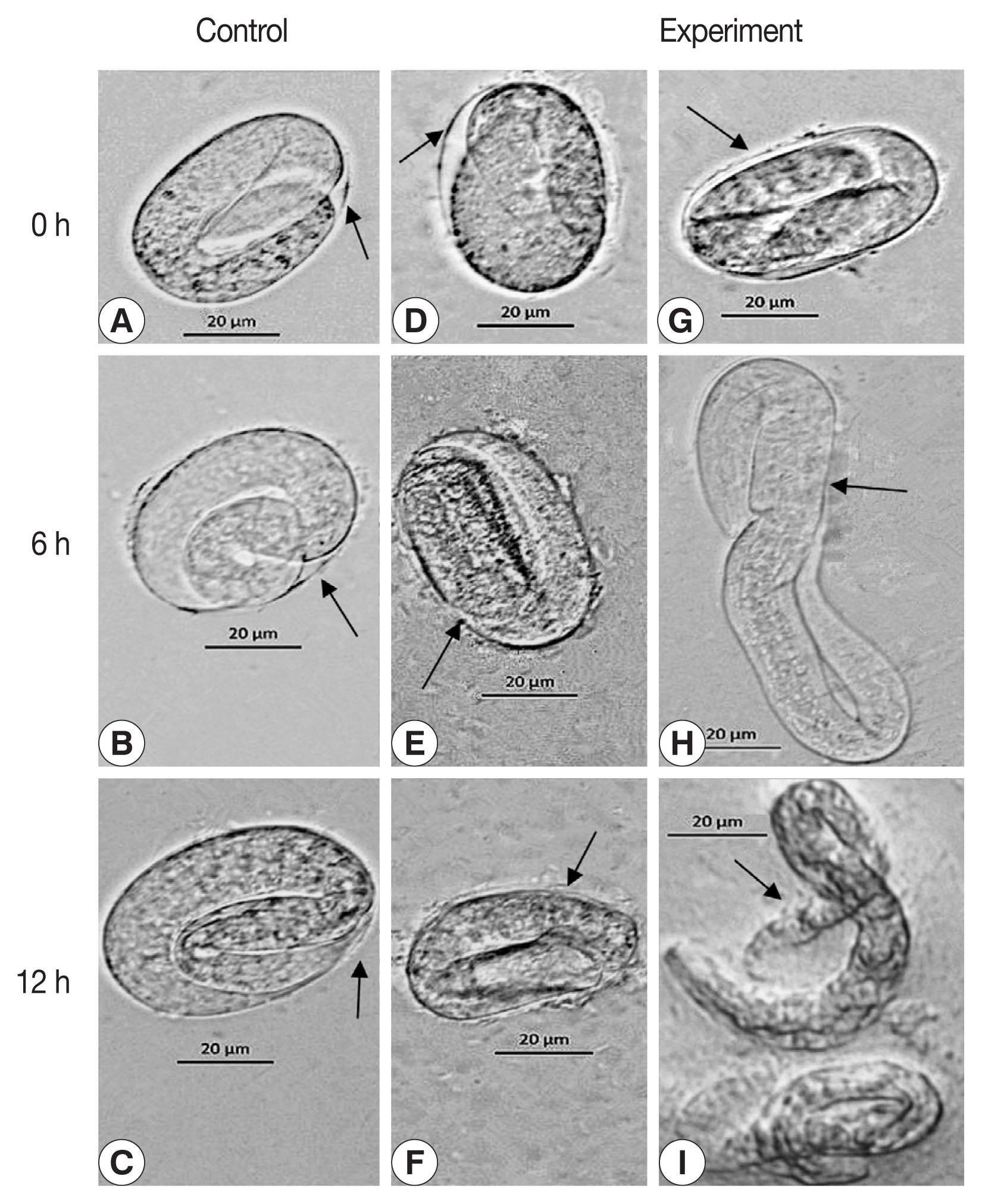
The degradation role of reAO-801 against C. elegans eggs. Control: (A–C) C. elegans eggs treated with the inactivated reAO-801 after 0, 6, and 12 h, respectively. Experiment: (D–F, G–I) C. elegans eggs treated with the reAO-801 after 0, 6, and 12 h, respectively.
DISCUSSION
Most of the chitinases of nematophagous fungi belong to the GH18 family [6,7,17,19,20]. The typical GH18 family of chitinases generally contains a signal peptide, a chitinase catalytic domain (CD), a chitinase binding domain (BD), and a short C-terminal structure, and some chitinases also contain a chitinase insertion domain (CID) [7,21–26]. Yang et al. [8] identified 16 chitinases in A. oligospora with some differences in the predicted molecular structure, confirming that different chitinases might have some function and expression level during nematode predation. Our molecular characterization of AO-801 revealed that it belongs to the GH18 family and had a CID, a substrate-binding site SIGGW, and a hydrolase catalytic active site LDGVDVDWE. The AO-801 of Arthrospora oligospora Xinjiang isolate A1 (A. oligospora XJ-A1) exhibited great similarity with the standard strain (ATCC 24927), with an amino acid sequence homology of 97.34%. Both have a typical (α/β)8 TIM barrel structure, and their amino acid sequences in the conserved regions are completely identical, especially the SXGG in the substrate-binding site and the active catalytic site of DXXDXDXE. Asp121, Asp124, Asp126, and Glu128 in the catalytic domain of fungal chitinases are essential, especially Glu is considered to be the proton acceptor of its active center [8,13,21,22]. In addition, the interaction of CID with the TIM barrel results in the strong binding to the substrate, which facilitates the degradation of the substrate. Li et al. [25] aligned 27 chitinase sequences containing chitinase insertion domains across taxa and demonstrated that the TIM barrel domain and CID could bind long-chain substrates by providing a deep substrate-binding cleft. In addition, there are a large number of hydrophobic amino acids in the amino acid sequence of AO-801, which is beneficial to maintaining the (α/β)8 barrel structure.
Yang et al. [8] performed a phylogenetic analysis of 16 chitinases from diverse nematophagous fungi. The 16 chitinases from A. oligospora were grouped into 4 subclasses, among which chitinases AO-801, AO-31, and AO-587 were clustered into one evolutionary branch, and were more closely related to the chitinases of phytopathogenic fungus Magnaporthe oryzae [8]. In the present study, XJ-A1 AO-801 was homologous to the nematophagous fungal chitinases of D. cylindrospora (KAF3934333) and D. brochopaga (KAF3910283), and also showed a closer relationship with the chitinase of plant pathogenic fungi.
Chitinases degrade chitin by catalyzing the hydrolysis of β-1, 4 glycosidic bonds of long-chain chitin to glucosamine monomers [27]. Most nematode eggshells are composed of protein and chitin. Chitin constitutes about 40% of the eggshell and is the main barrier to preventing infection. It is also essential for egg development and affects fertilization and polarization [14,28,29]. The mechanism of nematode predation by fungi is through enzymatic action to soften and disintegrate the host chitin wall [30]. The funcgal chitinases treated to Meloidogyne hapla eggs and juveniles cause premature hatching of nematode eggs, leading to the juveniles death [31]. The chitinase Chi43 from P. rubescens and P. chlamydosporia exhibited significant degradation activity against G. pallida eggs [15]. The chitinase of L. antillanum B-3 degraded or softened the eggshell and interfered with hatching of M. incongnita eggs [17]. In the present study, we confirmed that the recombinant reAO-801 killed stage-I larvae of C. elegans and degraded eggs of C. elegans and H. contortus. The chitinase AO-801 degraded C. elegans eggshells leading to premature hatching and subsequent death of the nematode eggs. This phenomenon was consistent with the previous results [15,30,31]. The chitinase AO-801 plays a vital role in the nematode predation. Yang et al. [8]found that the expression of chitinase AO-801 of A. oligospora increased by 2.75 times under the induction culture of cell walls of pathogenic fungus Rhizoctonia solani containing chitin components. They also found that most of the chitinases in A. oligospora had increased expression levels in response to nematode stimulation or environmental stress, suggesting that chitinase might be involved in the biotic or abiotic stress responses of A. oligospora. However, the action mechanism of chitinases during nematode infestation by nematophagous fungi requires further investigation.
In conclusion, this study elucidated the molecular characteristics of chitinase XJ-A1 AO-801 and obtained the enzymatically active reAO-801 for the first time through heterologous expression of AO-801 in P. pastoris. Our study confirmed that reAO-801 could kill stage-I larvae of C. elegans and degrade C. elegans and H. contortus eggs, and had strong nematicidal activity. Our findings provide an important foundation for the development of nematophagous fungus-based biocontrol strategies.
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (32060801). We thank the staff who provided the technical assistance for this study.
Notes
The authors declare that they have no conflict of interest.