Phylogenetic and phylogeographic analyses of Anisakis simplex sensu stricto (Nematoda: Anisakidae) from the common minke whale in Korean waters
Article information
Abstract
The genus Anisakis is among the most significant parasites to public health, as it causes anisakiasis, a parasitic infection in humans resulting from consuming raw or undercooked seafood. Although the infection status of Anisakis in second intermediate hosts, such as marine fishes and cephalopods, and humans have been severally reported in Korea, no information about the definitive host in Korean waters is available. In 2014, 2 adult gastric nematodes were collected from a common minke whale (Balaenoptera acutorostrata) found in the East Sea, Korea. These worms were identified as A. simplex sensu stricto (s.s.) by comparing the mitochondrial COX2 marker with previously deposited sequences. Phylogenetic and phylogeographic analyses of A. simplex (s.s.) worldwide revealed 2 distinct populations: the Pacific population and the European waters population. This is the first report on adult Anisakis and its definitive host species in Korea. Further studies on Anisakis infection in other cetacean species and marine mammals in Korean seas are warranted.
Introduction
The genus Anisakis comprises a well-known group of nematodes that pose a significant public health risk by invading the gastrointestinal walls of humans. The life cycle of this genus involves crustaceans as the first intermediate host, fish and cephalopods as the second intermediate/paratenic host, and marine mammals as the definitive host [1]. Based on morphological and genetic characteristics, the genus is divided into 2 groups: Anisakis type 1 (including A. simplex sensu stricto (s.s.), A. pegreffii, A. berlandi, A. typica, A. nascettii, and A. ziphidarum) and Anisakis type 2 (including A. physeteris, A. brevispiculata, and A. paggiae) [1]. Anisakiasis, caused by accidental ingestion of Anisakis larvae through raw or undercooked seafood, leads to symptoms such as epigastric pain, nausea, vomiting, diarrhea, allergic reactions (e.g., anaphylaxis), hemoptysis, hematemesis, intussusception, leukocytosis, and eosinophilia [2]. Anisakiasis cases have been reported worldwide, particularly in countries where eating raw or undercooked seafood is common, including Japan, the Netherlands, Germany, France, Spain, and Korea [2]. While Anisakis was previously considered the sole cause of infection, referred to as anisakiasis or anisakiosis, it has been discovered that Pseudoterranova and Contracaecum can also cause the disease. Thus, the term “anisakidosis,” using the name of the family, has recently been suggested [2].
Anisakiasis is one of the most important parasitic diseases causing public health concerns in Korea, with over 660 cases reported since 1971 [3–5]. However, the ecology of Anisakis in Korean waters is not well known. Research on Anisakis infection in fish and cephalopods, the second intermediate hosts directly responsible for anisakiasis, has been conducted consistently. Three Anisakis species, i.e., A. simplex (s.s.), A. pegreffii, A. typica, and a hybrid between A. simplex (s.s.) and A. pegreffii, have been identified at the species level in fish and cephalopods in Korea [6–11]. Until the early 2010s, when only morphological identification was available, it was difficult to identify these nematodes at the species level due to a lack of clarity on the taxonomical significance of this group [1]. Consequently, most Anisakis larvae were conventionally referred to as A. simplex type 1 or as A. simplex in Korea [12]. Subsequently, genetic diagnosis was recognized as a reliable identification method and extensively adopted [1]. In Korea, species level identification was also conducted, resulting in the recognition and reporting of other Anisakis species.
Eight species, namely, A. simplex (s.s.), A. physeteris, A. pegreffii, P. decipiens (s.s.), P. azarasi, P. cattani, C. osculatum, and Hysterothylacium aduncum have been reported as causative agents of human anisakiasis worldwide [2,13–15]. Among them, 3 species of Anisakidae larvae (A. pegreffii, A. simplex (s.s.), and P. decipiens (s.s.)) have been reported in Korea [2, 5,14,16]. The presence of larvae in intermediate hosts indicates the existence of definitive hosts harboring and transmitting this parasite in the ecosystem. However, there is currently no available information on the definitive host in Korea. To develop preventive measures for human anisakiasis, a major public health issue under the concept of “One Health,” it is necessary to elucidate the life cycle of anisakids in Korean waters. Therefore, studies on the infection status of definitive hosts are necessary. Identifying the definitive host species of Anisakidae and evaluating the relationship between the habitat range of the final host and the infection status of the intermediate host in the sea area are essential for establishing preventive measures against anisakiasis.
The common minke whale, Balaenoptera acutorostrata Lacépède 1804, is divided into 3 subspecies: the North Atlantic minke whale (B. a. acutorostrata), the North Pacific minke whale (B. a. scammoni), and an unnamed dwarf minke whale distributed in the Southern Hemisphere, depending on their habitat [17]. Balaenoptera a. scammoni is further classified into 3 populations based on comparisons of mtDNA and the white bands on pectoral fins; a population habiting in the East Sea-the Yellow Sea-the East China Sea, a population in the Sea of Okhotsk-the Northwest Pacific Ocean, and a population inhabiting east part of the North Pacific [18]. The Korean population belongs to the first group. Since the International Whaling Commission passed an official moratorium on commercial whaling in 1986, banning this activity, this species has been evaluated as having a sufficient population compared to other large baleen whales and is classified as “Least Concern” on the International Union for Conservation of Nature Red List [19]. In Korean waters, an average of 73 of these whales are bycaught annually [20], and they are not designated as domestic marine protection creatures. Thus, this species is still distributed on the market and consumed as whale meat. Despite the significant number of strandings or bycatch incidents, few investigations have been conducted on the infectious agents of this species due to limited research opportunities. Identifying anisakids at the species level is essential for understanding the distribution and epidemiology of common minke whales worldwide [21]. Therefore, we aim to (i) identify the species of adult anisakid worms detected in the definitive host and (ii) analyze the phylogenetic relationships and population genetic structure of the identified Anisakis species using previously deposited sequences in GenBank.
Materials and Methods
Anisakis worm from the common minke whale
A 4.5-m-long common minke whale (B. a. scammoni) careass was found in the sea between Namhae, Gyeongsangnam-do, and Bangeojin, Ulsan, Korea, and dissected in 2012 (Supplementary Fig. S1). During the disassembly, the gastrointestinal tract was exposed, and 2 adult roundworms were detected.
Polymerase chain reaction
The 2 adult worms were frozen and then fixed in 70% ethanol, which resulted in decomposition, making it unsuitable to observe and identify their detailed organs morphologically. Therefore, DNA from the 2 specimens were extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). The partial mitochondrial cytochrome oxidase 2 (COX2) markers, which has been analyzed the most in Anisakidae, was amplified using the polymerase chain reaction (PCR) method. The primers used for amplification were as follows: forward: 211 (TTTTCTAGTTATATAGATTGRTTYAT) and reverse: 210 (CACCAACTCTTA AAATTATC) [S1]. The COX2 region was amplified using 2 μM of template DNA, 6 μl of 5×PCR Master Mix (Elpis Biotech, Daejeon, Korea), and 7.5 pmol of each primer in a final volume of 30 μl. The PCR amplification conditions were as follows: 3 min at 94°C (initial denaturation), 35 cycles of 30 sec at 94°C (denaturation), 60 sec at 46°C (annealing), and 90 sec at 72°C (extension), with a final elongation step of 10 min at 72°C.
Phylogenetic analyses
The PCR products were sequenced by Cosmogenetech (Seoul, Korea). Consensus sequences were assembled using Geneious ver. 2022.2.2. [S2] and compared with previously registered sequences on GenBank using the basic local alignment search tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) for species identification [S3]. Phylogenetic analysis was conducted on 576 base pairs (bp) of the COX2 marker using previously deposited sequences of the same species detected from intermediate and definitive hosts, as well as sequences of other species in the same genus. In the case of the same species, sequences from Korean seas and definitive hosts in other waters were all included if the sequence lengths were adequate for analysis. Among foreign sequences of larvae, if several sequences were registered by 1 study, we included only 1 sequence with the highest correspondence to our sequence. One sequence was used as a representative of other Anisakis species. The sequences used in the phylogenetic analysis are shown in Supplementary Table S1. We included sequences from the Northwest Pacific Ocean (n=42, all isolated from intermediate hosts), the Northeast Pacific Ocean (n=2, all isolated from definitive hosts), the Northeast Atlantic Ocean (n=14, 12 isolated from definitive hosts, 2 isolated from intermediate hosts), the Mediterranean Sea (n=6, all isolated from intermediate hosts), and the North Sea (n=6, all isolated from intermediate hosts).
The COX2 sequences were aligned using Clustal W with Mega X software [S4]. To reconstruct the phylogenetic tree, the best-fit substitution model of sequence evolution was selected using Jmodeltest 2.1.10 [S5], which was implemented based on the Bayesian information criterion. The best-fit model, GTR+I+G, and its parameters were used in the analysis. Bayesian inference (BI) tree was constructed using MrBayes 3.2.6, with 10,000,000 generations and the default value of 4 Markov chains [S6]. Maximum likelihood (ML) tree was constructed using the IQTREE web server (http://iqtree.cibiv.univie.ac.at) [S7]. The phylogenetic trees were rooted using C. rudolphii as outgroup and then concatenated. The final trees were visualized using FigTree v.1.4.4. [S8].
Phylogeographic analyses
For population genetic analysis, all sequences of A. simplex (s.s.) used in the phylogenetic comparison were included, except for 3 sequences where the collection sea area was not clearly specified in either the publication or GenBank (AP017678, DQ116426, and MN877346). The evolutionary relationships of the COX2 regions were visualized using a median-joining network based on NETWORK ver. 10.2 software (https://www.fluxus-engineering.com) [S9].
Results
The lengths (90.0 mm and 91.5 mm) and widths (3.5 mm and 3.55 mm) of the 2 worms were measured (Fig. 1A). Both worms were female and had eggs inside the uterus. The average diameter of randomly selected eggs was 49.98±2.83 μm (n=20) (Fig. 1B).

Adult worms and an egg of Anisakis simplex sensu stricto (s.s.) recovered from the common minke whale, Balaenoptera acutorostrata. (A) Two female adult Anisakis simplex (s.s.). (B) The egg of Anisakis simplex (s.s.) observed under a light microscope.
The 2 specimens showed the same haplotype of the COX2 marker. As a result of the direct sequencing of the partial COX2 region, the adult nematodes recovered from the common minke whale were identified as A. simplex (s.s.). The sequence showed high concordance (97.4–99.8%) with previously deposited sequences of the same species larvae detected in second intermediate hosts in Korean seas. Additionally, it was confirmed to have an identical haplotype with a sequence (LC543749) from Skipjack tuna (Katsuwonus pelamis) in Japanese waters [S17]. The newly generated sequence was registered in GenBank with accession number OP948879.
Based on the BI and ML phylogenetic analyses using the mtDNA COX2 marker, the genus Anisakis was divided into 2 main clades (Fig. 2). The first group included 6 species of Anisakis type 1: A. simplex (s.s.), A. pegreffii, A. berlandi, A. typica, A. nascettii, and A. ziphidarum. The second group clustered 3 Anisakis type 2 species: A. physeteris, A. brevispiculata, and A. paggiae. Among the 9 species of the genus Anisakis, a group of 72 sequences of A. simplex (s.s.), including our sequence, was the most recent to diverge. Within these 72 sequences, intraspecies relationships showed 2 distinct clades based on sea area. The Pacific clade comprised 38 sequences from the Northwest Pacific Ocean and 2 from the Northeast Pacific Ocean, and it was found to be paraphyletic to the European clade, which included sequences from the Northeast Atlantic Ocean, the Mediterranean Sea, and the North Sea, although with a lack of significant statistical support. Two exceptions from the Northwest Pacific Ocean were observed. One sequence (AY994157), registered as A. simplex (s.s.), clustered with a reference sequence of A. pegreffii (DQ116428) with significant node values (1/86). Another sequence (AP017678), deposited by a Japanese research group without citing the sea origin of the Anisakis larvae, clustered within the European clade. The intraspecific haplotype diversity of the A. simplex (s.s.) group was 0.996.
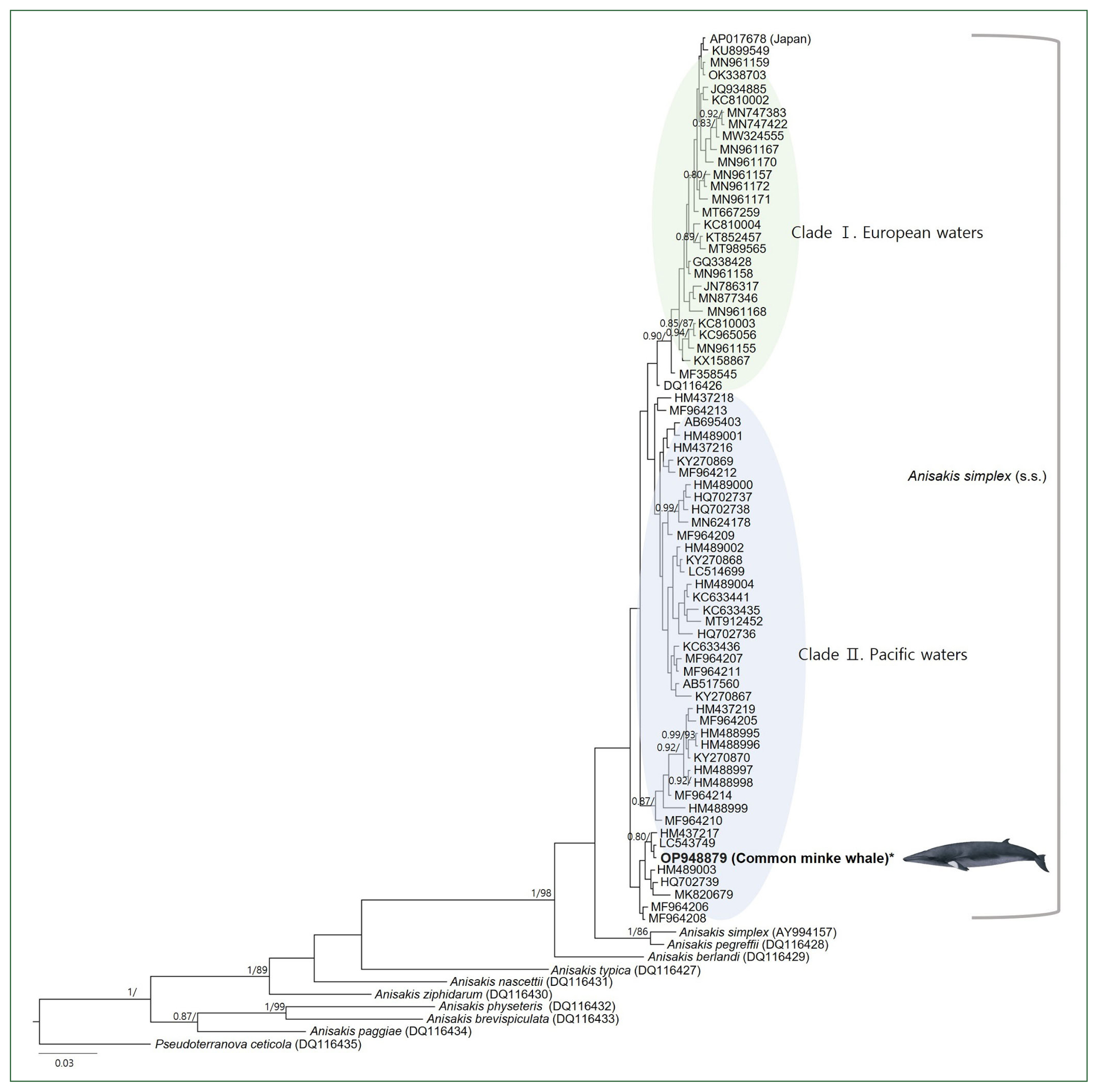
A concatenated phylogenetic tree from Bayesian inference (BI) and max likelihood (ML) analyses of the mitochondrial cytochrome oxidase 2 (COX2) datasets constructed based on the GTR+I+G model of evolution. The newly generated sequence in this study is indicated in bold. Posterior probability values (≥0.80) and bootstrap values (≥85) are given on the branches. The branch length scale bars indicate the number of substitutions per site. According to the BI and ML phylogenetic analyses using the COX2 marker, the Anisakis simplex (s.s.) sequences formed 2 highly supported clades.
Population genetics inferred from the COX2 marker also showed grouping divided by the Pacific and European sea areas due to excluding sequences without a clearly specified origin (Fig. 3). Two haplotypes in European waters were shared by specimens from the Northeast Atlantic Ocean and the North Sea. Most specimens from the Northwest Pacific had unique haplotypes, except for 3 haplotypes, which were each shared by 2 individuals.

Genealogical relationships among 69 mitochondrial COX2 marker haplotypes of Anisakis simplex (s.s.) from 5 different sea areas inferred from all available GenBank sequences based on a median-joining (MJ) network. The diameter of each circle is proportional to the number of specimens. The short vertical lines indicate single mutational differences between the 2 haplotypes connected. Circle colors represent geographical origins from which particular anisakid haplotypes were sampled.
Discussion
A. simplex (s.s.) is distributed in seas worldwide and is commonly found in the North Atlantic and North Pacific Oceans, where it overlaps with A. pegreffii [22]. The morphological characteristics of these 2 species are closely similar and challenging to distinguish, making genetic identification essential [1]. A mitochondrial full sequence of A. simplex (s.l.) (AY994157), which was examined in Korea [23], clustered with a reference sequence of A. pegreffii (DQ 116428) in the phylogenetic analysis. This finding aligns with a previous study that analyzed the mitochondrial full genome [24]. The degree of identity with our newly generated A. simplex (s.s.) COX2 sequence was 95.4%. Considering these results, this could be an A. pegreffii larva, which was difficult to distinguish morphologically, especially in 2006, when A. pegreffii was classified as Anisakis type 1 without much knowledge. Our specimens were confirmed as A. simplex (s.s.) by showing over 97.4% concordance with all other A. simplex (s.s.) sequences collected in Korean seas, and the phylogenetic analysis supported this result.
One COX2 sequence (AP017678), deposited as A. simplex (s.s.) by a Japanese research team, was excluded from the population genetics analysis because the collection sea area of the specimen was not clearly defined and because of the phylogenetic analysis result. This sequence showed an identical haplotype to those in the Baltic Sea (OK338703) [S24] and the Northeast Atlantic Ocean (MN961159) [S21], and only a single bp difference from a haplotype in the Baltic Sea (KU899549) [S15]. In the phylogenetic analysis, the tree was largely divided into 2 groups, namely, the Pacific clade and the European clade (including the Northeast Atlantic Ocean, the Mediterranean, and the North Sea), except for AP017678. This exempted sequence has not yet been published; thus, the species and origin of the host are unknown. Based on this phylogenetic result, the parasite’s host could be fish or cephalopods imported from European waters to Japan.
Nine Anisakis species, including the A. simplex (s.s.) group, were divided into 2 monophyletic clades (Anisakis type 1 and type 2), consistent with previous studies. The diverging order of these 9 species was also consistent with previous surveys [21]. For the A. simplex (s.s.) group, sequences from European waters formed 1 clade, while Pacific sequences were largely divided into 3 clades and grouped as paraphyletic to the European clade. This result is consistent with the phylogeographic analysis, which also divided the haplotypes into European and Pacific groups. In other words, the distinct populated genetic structure based on the sea area appeared to be recognized. The divided regional distribution and genetic structure of host species could contribute to maintaining genetic differentiation among the parasites [25]. Conversely, the structure was not significantly populated in the Pacific Ocean or in European waters. This indicates that mitochondrial gene flow within the same sea area is high, possibly because definitive cetacean hosts or second intermediate hosts preying or breeding play a role in moving parasites [25]. However, the limited number of sequences from certain sea areas, such as the Northeast Pacific Ocean, the Mediterranean Sea, and the North Sea, hindered more accurate analyses, highlighting the need for a more extensive examination by adding sufficient sequences from these waters.
Since the 2000s, there have been several reports on the infection status of Anisakis in second intermediate hosts, such as fish and cephalopods, in Korea [6–11]. The adult worms we found in the common minke whale were identified as A. simplex (s.s.), which had been recognized as a main causative agent of anisakiasis and a major Anisakis species in second intermediate hosts in Korea [12]. However, using molecular techniques, recent studies have shown that most nematodes found in second intermediate hosts in Korean waters and in Korean patients are A. pegreffii [5,9,10,14]. Therefore, further research on definitive hosts and the distribution of A. pegreffii is required.
Verifying the origin waters of intermediate hosts is essential for the epidemiological investigation of human anisakiasis [10]. The distribution of anisakids in Korea varies depending on the sea area and fish species. For example, most fish and cephalopod species in the Yellow Sea, Southern Sea, and East Sea were found to be predominantly infected with A. pegreffii [8–10], while 2 salmonid species, 1 gadoid fish, and common squid collected in the East Sea were mainly infected with A. simplex (s.s.) [6–9,11]. This difference may be due to factors such as water temperature, preying sources, and topographical characteristics. Moreover, the distribution and movement of definitive hosts for each anisakid species could also play a role [10]. Analyzing the distribution of second intermediate host species and their Anisakis species in connection with the distribution area of definitive hosts in Korean seas is crucial for devising preventive measures for human anisakiasis.
The common minke whale inhabits the East Sea, the Yellow Sea, and the eastern part of the Southern Sea of Korea, migrating according to the season [26]. The East Sea population lives in waters between 13–16°C and migrates based on changes in water temperature, staying in the Southern Sea in winter, moving north in spring, and migrating south again in autumn and winter [26,27]. A. simplex (s.s.) collected from second intermediate hosts in Korean waters has mainly been found in the East Sea [6–10]. Considering the wide range of travel of the common minke whale in the East Sea, depending on the season, they may act as definitive hosts to maintain the life cycle of A. simplex (s.s.) in this sea area.
The genus Anisakis shows a definitive host preference based on geographical distribution, including sea depth, the preying ecology of definitive hosts, and the coevolutionary history between definitive hosts and parasites [1]. The definitive host species of A. simplex (s.s.) reported so far include the common minke whale (B. acutorostrata), common dolphin (Delphinus delphis), pilot whale (Globicephala melaena), white-beaked dolphin (Lagenorhynchus albirostris), killer whale (Orcinus orca), and striped dolphin (Stenella coeruleoalba) [28]. Among these species, common dolphins and killer whales, which reportedly inhabit Korean waters, especially the East Sea, may also play a role in maintaining the life cycle of A. simplex (s.s.) in this sea area, along with the common minke whale. Therefore, investigating the infection status of other cetacean species that could potentially act as definitive hosts for Anisakis is necessary to accurately reveal the distribution and host range of anisakids in Korean seas. Additionally, examining Anisakis infection in the common minke whale population in the Yellow Sea is important since only A. pegreffii, not A. simplex (s.s.), has been reported from second intermediate hosts in the Yellow Sea [8–10]. Long-term monitoring of host infection status is also necessary because climate change may directly affect the free-living stage of anisakids and indirectly affect the second intermediate and definitive hosts, resulting in differences in the prevalence and degree of infection [10].
Conversely, parasite composition and infection degree can serve as biological tags to estimate the migration pathways and population stocks of migratory host species [29–31]. Anisakis, in particular, is a useful taxon as a biological tag because it can survive relatively long in the host, and there is a significant amount of knowledge about its geographical distribution and life cycle compared to other groups [32]. The population genetic structure of definitive hosts can be inversely estimated from the population genetic structure and phylogeography of Anisakis species, allowing for co-phylogeographical analysis based on this information [21]. In other words, it is possible to estimate and analyze ecological information, such as the distribution, movement, population, and food habits of whale species, by investigating parasite infections. It was previously assumed that the common minke whale populations in Korean waters were not divided between the East Sea and the Yellow Sea [33]; however, this conclusion was reached before genetic analysis was available. Presently, there is a possibility of 2 or more stocks, similar to the waters of neighboring Japan [34], but this has not yet been studied. Since the genetic substructure of endoparasites overlaps with the population of definitive hosts [25], parasites can be used to analyze the population of the common minke whale in Korean waters. To analyze the genetic structure, phylogeography, and ecology of hosts and parasites, a combined approach is essential.
This is the first report on adult Anisakis in Korean waters. Furthermore, this is the first study examining Anisakis species in the common minke whale inhabiting the East Sea. A previous study examined the host species in Japanese waters, but only Japan’s eastern and northern coasts were included [30]. Proper necropsy of marine mammals allows for intact parasite sampling. However, for common minke whales sold in Korean markets, a complete necropsy is nearly impossible, and parasites can only be obtained accidentally during disassembly, as in this case. A. simplex (s.s.) and A. pegreffii are found together in the same host individuals in their sympatric waters [25,35]. Syntopic infection and even hybrids of the 2 species have been confirmed in common minke whales in adjacent Japanese waters, with an average intensity of several thousand parasites [30]. In the present study, examining the accurate parasite infection status of the host was impossible, since a proper necropsy could not be performed and only 2 adult worms were obtained. Based on previous studies and the infection status of A. pegreffii in other definitive host species in Korean waters (unpublished data), we hypothesize that common minke whales in Korean waters can also be infected with adult A. pegreffii. We strongly recommend sampling intact parasites through proper necropsy of various cetacean species to investigate the infection status of Anisakis clearly and synthetically in the definitive hosts and describe their morphological characteristics. It will be helpful to establish appropriate precautionary measures against human anisakiasis in Korea.
Supplementary Information
Acknowledgement
This research was supported by Chungbuk National University: Korea National University Development Project (2020).
Notes
Author contributions
Conceptualization: Kim S, Choe S
Data curation: Kim S, Choe S
Formal analysis: Kim S, Choe S
Funding acquisition: Choe S
Investigation: Kim S, Lee BS, Choe S
Methodology: Kim S, Choe S
Project administration: Kim S, Choe S
Supervision: Choe S
Visualization: Kim S
Writing – original draft: Kim S
Writing – review & editing: Kim S, Choe S
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
