INTRODUCTION
Human sparganosis is a zoonotic disease caused by infection with the larval forms (procercoid/plerocercoid) of
Spirometra spp. The first recorded case of human sparganosis in Korea involved a farmer [
1] based on a report by Uemura in 1917. Cho et al. [
2] reviewed 63 cases of indigenous sparganosis in 1975 in Korea. Then, an additional 56 cases were presented during the period from 1975 to 1989 [
3]. The most recent report published in 2015 details human sparganosis cases recorded for the period 1979 and 2009 in Korea involved
Spirometra erinaceieuropaei (Rudolphi, 1819) (n=35) and
Spirometra decipiens (Diesing, 1850) (n=15) [
4]. As a consequence,
S. decipiens was added to the causative agent list of human sparganosis that was previously regarded as only involving
S. erinaceieuropaei.
The routes of spargana infection involve either drinking water contaminated with procercoid-infected copepods or consumption of undercooked meat of plerocercoid-infected snakes or frogs. Additionally, infection can occur by placement of poultices that use the skin of infected snakes or frogs. Snakes have been noted as a very important source of infection for human sparganosis by many researchers in Korea [
5-
7]. At least 7 species of terrestrial snakes have thus far been investigated as possible sources of infections with spargana, among which the following 6 species of snakes have been positively identified with spargana:
Elaphe rufodorsata,
Elaphe dione,
Dinodon rufozonatum ruforzonatum,
Rhabdophis tigrinus tigrinus,
Zamenis spinalis, and
Agkistrodon halys. All of these snakes were from Gangwon-do (Province), which is located at the eastern side of the peninsula [
8]. Spargana have been found in various species of frogs, snakes, birds, and mammals in the Far East region, including China and Japan, and 6 spirometrid species (under the name
Diphyllobothrium) were reported with morphological and biological studies on the species in China [
9]. The present study was to provide information for what
Spirometra species are being distributed in Korea and China by using genetic analysis.
The morphological identification of spargana is usually difficult because they lack distinguishing characteristics. More recently, molecular approaches to differential identification of
S. erinaceieuropaei and
S. decipiens have been introduced that analyze sequence differences of mitochondrial genomes [
4,
10]. In the present study, we used species-specific primers for a multiplex PCR assay based on mitochondrial sequences of
S. erinaceieuropaei and
S. decipiens. The larval forms of
Spirometra spp. (spargana) collected from terrestrial snakes in Korea and China were identified by the multiplex PCR assay and through phylogenetic analysis of mitochondrial DNA sequence data.
DISCUSSION
It is known that the majority of human sparganosis cases are caused by consumption of raw reptiles (snakes), amphibians (frogs), or drinking water contaminated with larval
Spirometra species. Regarding the well-known species
Spirometra erinaceieuropaei, Cho et al. [
2] investigated 56 cases in search of possible sources of infections for human sparganosis. Approximately a half of the cases (n=30) were involved with consumption of raw snakes, and 10 cases were associated with drinking untreated water. Thus, the consumption of raw snake flesh has been considered the most important cause of human sparganosis in Korea. This was generally accepted by most parasitologists without opposition; however, our research group recently suggested that
S. erinaceieuropaei is not the only species that induces human sparganosis [
4]. The suggestion was that
S. decipiens is another possible cause of human sparganosis in Korea. The species ratio presented in the report was 35 (
S. erinaceieuropaei): 15 (
S. decipiens) for cases of human sparganosis among Korean patients recorded for the period 1979-2009 from all of the parasitology laboratories in Korea. The simplified ratio of 2.3:1 reveals approximately twice as many cases of
S. erinaceieuropaei than
S. decipiens among the 50 specimens examined [
4]. This finding immediately provoked questions concerning whether
S. erinaceieuropaei and
S. decipiens are caused by the same source of infection, namely, snakes, and if so, what is the ratio between these
Spirometra species in the snakes? To answer these questions, genetic analyses of spargana that infect snakes are required to identify the
Spirometra species. Our present study revealed a rather surprising result that all spargana from snakes were identified as
S. decipiens (n=904) without exception. This finding highlights a serious epidemiological discrepancy between the ratios of
Spirometra species in humans and snakes.
We do not currently know the reason why we did not find any
S. erinaceieuropaei from any of our snakes. One possible explanation is that the
S. erinaceieuropaei larval stage may be the primary source of human infection by way of drinking water contaminated with procercoids, but this then begs an even bigger question: what is the intermediate host that transfers larval forms to humans if it is not snakes? Furthermore, what is the final host? However, all these questions are based on a supposition that there are 2 different kinds of
Spirometra in humans. The use of a poultice, which represents another form of spargana transmission to humans, was not taken seriously because it seems that it is not commonly practiced nowadays. The final hosts of
Spirometra spp. are commonly known to be carnivorous animals such as cats and dogs, but
S. erinaceieuropaei has not been found or ever described in natural infections in cats and dogs [
9]. Thus,
S. decipiens might be the only species of naturally occurring tapeworm that is currently found in cats and dogs. The only clearly known second intermediate host of
S. erinaceieuropaei is the Chinese hedgehog. These findings indicate that further studies are required to determine the real infection route and final hosts of
S. erinaceieuropaei, and to clarify whether
S. decipiens infects humans through drinking water contaminated with procercoids.
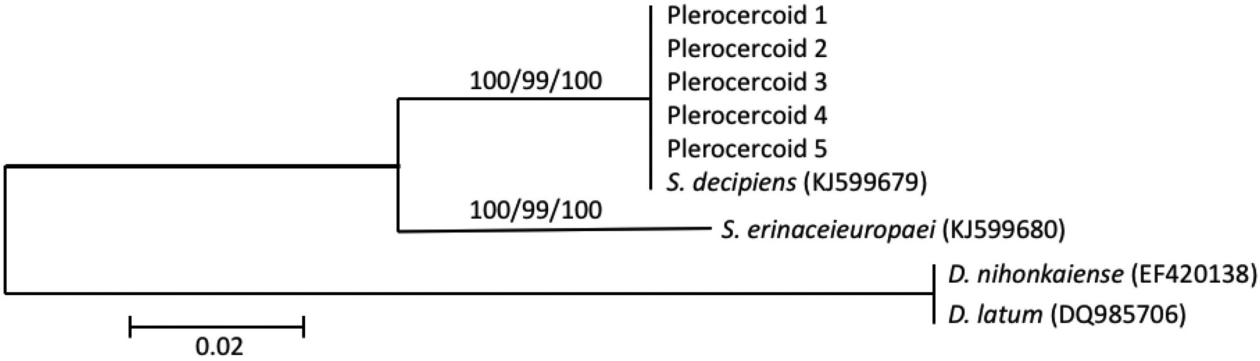
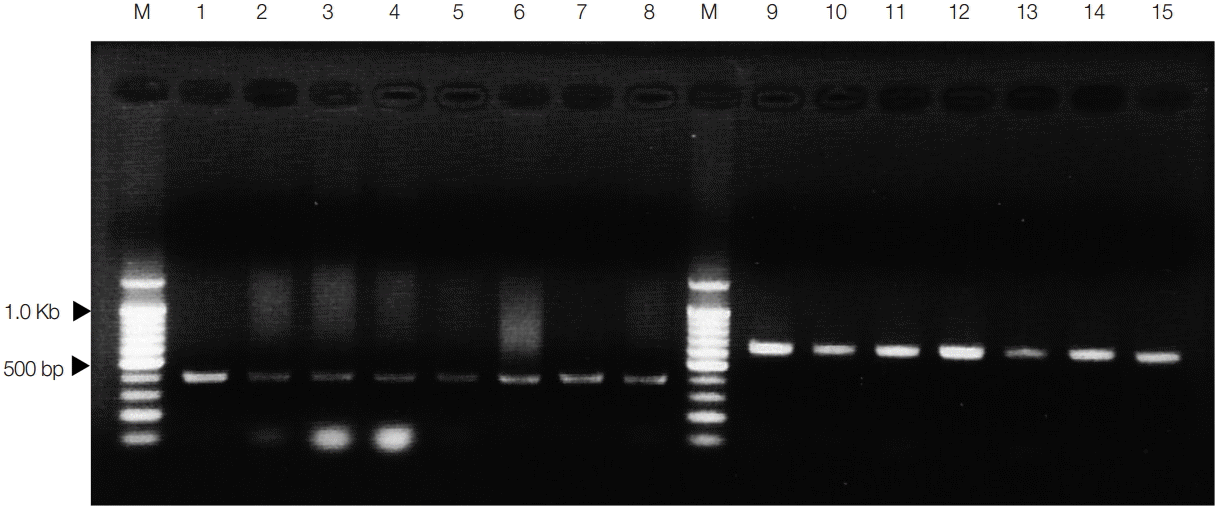
In the present study, the differential diagnosis obtained by PCR using species-specific primers was correlated with species identification based on nucleotide sequence analysis of the mitochondrial cox1 gene. The results showed that the multiplex PCR assay with the Se/Sd-7963F, Se-8344R, and Sd-8584R primers will be useful for species identification in the genus Spirometra. Finally, the spargana that infected snakes (D. rufozonatum rufozonatum, R. tigrinus tigrinus, and A. saxatilis) in Korea and China was S. decipiens, and this species was distinguished from the spargana of S. erinaceieuropaei using analysis of mitochondrial cox1 sequences and multiplex PCR. We wonder if our mysterious finding that S. erinaceieuropaei larval forms were not identified in snake specimens collected from Korea and China will be explained in the near future if investigations use larger sample sizes.