Introduction
Materials and Methods
In silico identification of SET-HKMT proteins
Phylogenetic analysis
Immunological detection of methylated histone derivatives
Temporal expression profiles of C. sinensis genes
Methyltransferase inhibitors and gene expressions
Results
Identification of SET-HKMTs in C. sinensis and other helminths
Table 1
| Accession No. | Source | Name | Length (aa) | Structural organization of PFAM domainsa | SET domain | ESTb |
|---|---|---|---|---|---|---|
| GAA49834c | GenBank | Histone-lysine N-methyltransferase SETDB | 2,189 |

|
Yes | + |
| GAA48712 | GenBank | Histone-lysine N-methyltransferase SETD8 | 340 |

|
Yes | + |
| GAA50403 | GenBank | Histone-lysine N-methyltransferase SETDB | 908*d |

|
No | − |
| GAA52264 | GenBank | Histone-lysine N-methyltransferase SETD1 | 128 |

|
No | − |
| GAA52032 | GenBank | Histone-lysine N-methyltransferase MLL5 | 892 |

|
Yes | + |
| GAA39455 | GenBank | Histone-lysine N-methyltransferase MLL3 | 1,443 |

|
Yes | + |
| GAA52505 | GenBank | Histone-lysine N-methyltransferase SETD2 | 757 |

|
Yes | + |
| GAA57198 | GenBank | Histone-lysine N-methyltransferase NSD2 | 1,293 |

|
Yes | + |
| GAA32467 | GenBank | Histone-lysine N-methyltransferase MLL3 | 1,763* |

|
Yes | + |
| GAA57433 | GenBank | Histone-lysine N-methyltransferase Trithorax | 328* |

|
Yes | + |
| GAA55675 | GenBank | Histone-lysine N-methyltransferase SUV39H2 | 436 |

|
Yes | + |
| GAA31138 | GenBank | Histone-lysine N-methyltransferase SETD1B | 1,685 |

|
Yes | + |
| GAA51760 | GenBank | Histone-lysine N-methyltransferase NSD1/2 | 1,596 |

|
Yes | + |
| GAA53884 | GenBank | Histone-lysine N-methyltransferase SUV420H | 527* |

|
Yes | + |
| GAA55462 | GenBank |
Histone-lysine N-methyltransferase Enhancer of Zeste |
940* |

|
Yes | + |
| GAA29510 | GenBank | Histone-lysine N-methyltransferase MLL5 | 973 |
|
Yes | + |
| GAA54150 | GenBank | Histone-lysine N-methyltransferase SETMAR | 262 |

|
Yes | + |
| GAA48681 | GenBank | Histone-lysine N-methyltransferase SETD2 | 1,000 |

|
No | + |
| GAA53888 | GenBank | SET and MYND Domain-containing Protein 4 | 817 |

|
Yes | + |
| GAA53887 | GenBank | SET and MYND Domain-containing Protein 4 | 869 |

|
Yes | + |
| GAA50167 | GenBank | SET and MYND Domain-containing Protein 4 | 761 |

|
Yes | + |
| GAA40653 | GenBank | SET and MYND Domain-containing Protein 5 | 575 |

|
Yes | + |
| GAA51355 | GenBank | SET Domain-containing Protein 4 | 493 |

|
Yes | + |
| GAA53907 | GenBank | Histone-lysine N-methyltransferase SETD3 | 254 |

|
No | + |
| GAA52155 | GenBank | Histone-lysine N-methyltransferase PRDM9 | 210 |

|
Yes | + |
| GAA36363 | GenBank | Enhancer of Zeste | 81 |

|
No | + |
| GAA50314 | GenBank | Histone-lysine N-methyltransferase MLL4 | 1,769 |

|
No | + |
| GAA28296 | GenBank | Histone-lysine N-methyltransferase SETD2 | 800 |

|
No | + |
| GAA53428 | GenBank | Histone-lysine N-methyltransferase MLL3 | 3,518* |

|
No | + |
| GAA50855 | GenBank | Histone-lysine N-methyltransferase MLL3 | 1,138 |

|
No | − |
| GAA52291 | GenBank | Histone-lysine N-methyltransferase ASH1L | 2,734 |

|
No | + |
| GAA47499 | GenBank | Histone-lysine N-methyltransferase PRDM9 | 1,031* |

|
No | + |
a The PFAM domains preserved in the amino acid sequeces of the Clonorchis SET-HKMT proteins were predicted by using SMART program (http://smart.embl.de/).
Structural characters of the helminth SET-HKMTs
 | Fig. 1Comparison of SET domain sequences conserved in the Clonorchis sinensis histone lysine methyltransferase (CsHKMT) proteins. The amino acid (aa) sequences of CsHKMT SET domains, which were defined in the full CsHKMT sequences using the SMART program, were aligned against one another for comparison of their primary structures. The degrees of the aa conservations are highlighted in different shades of grey, and gaps represented by dots were introduced in the alignment to increase the similarity values. The short aa signatures of the SET domain involved in the binding of S-adenosyl methionine (SAM) and histone Lys are presented above the alignment (Motifs I–IV). The solid triangle indicates the invariant Tyr forming H-bond/van der Waals interaction with the ε-amino group of histone Lys. Open arrowheads represent the positions of Gly, Asn, and His participating in the formation of the non-covalent bonds with SAM molecule. |
Phylogeny of the helminth SET-HKMTs
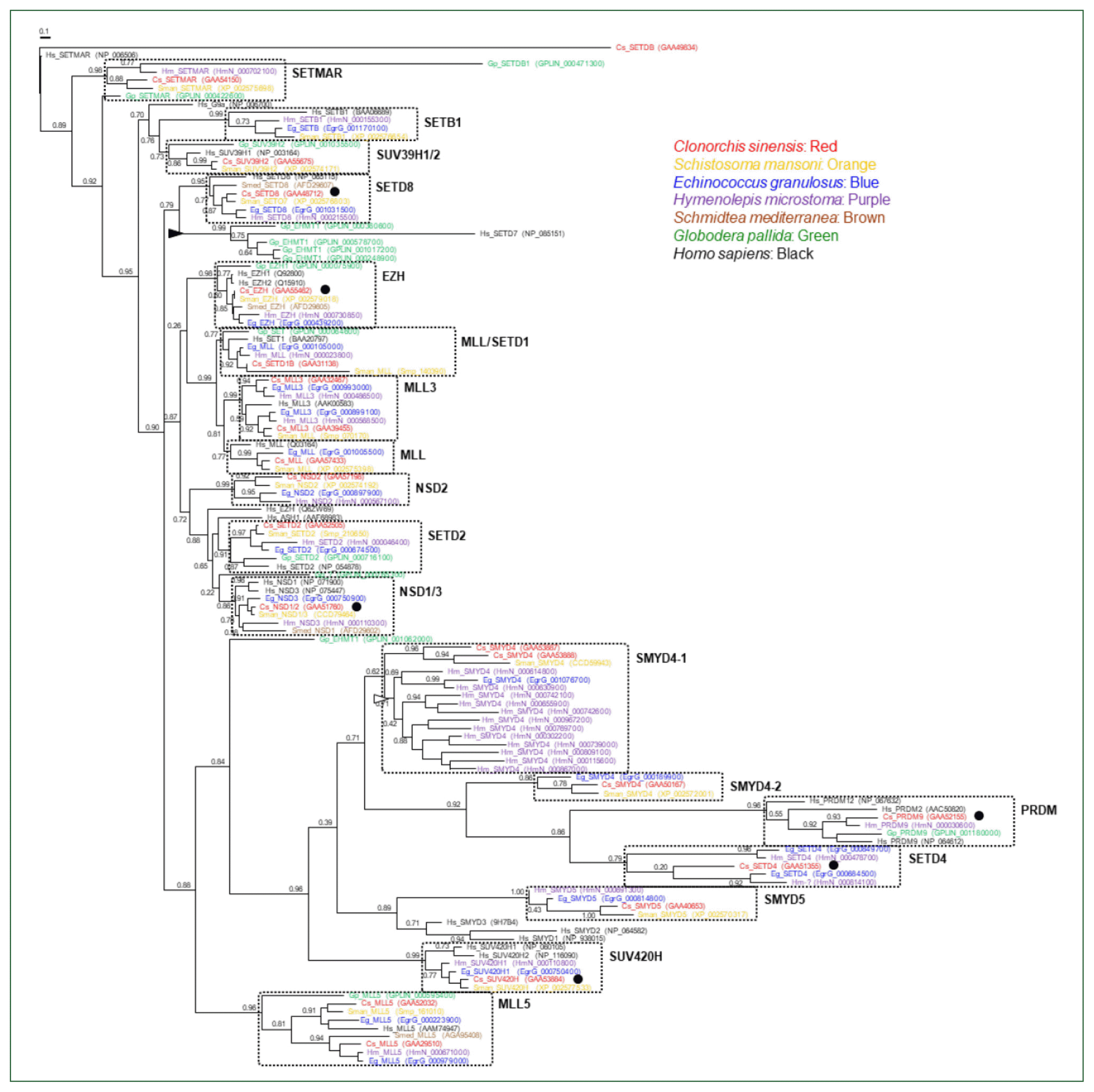 | Fig. 2Phylogeny of Clonorchis sinensis histone lysine methyltransferases (HKMTs) and their platyhelminth and nematode orthologs. The amino acid sequences of HKMT SET domains identified in trematodes (C. sinensis [red] and Schistosoma mansoni [orange]), cestodes (Echinococcus granulosus [blue] and Hymenolepis microstoma [purple]), turbellarian (Schmidtea mediterranea [brown]), nematode (Globodera pallida [green]), and human (black) were aligned and used in the phylogenetic analysis. The unrooted maximum likelihood tree was constructed using PhyML. Branch support values obtained using the Shimodaira-Hasegawa-like approximate likelihood ratio test are indicated at the corresponding branching nodes. Proteins, coding genes of which appeared to be expanded specifically in G. pallida and H. microstoma, are indicate with solid and open arrowheads, respectively. The solid circles indicate C. sinensis proteins selected for the gene expression analyses. |
Retrieving DOT1 protein sequences from the helminth proteomes
Comparative analysis of the helminth DOT1s
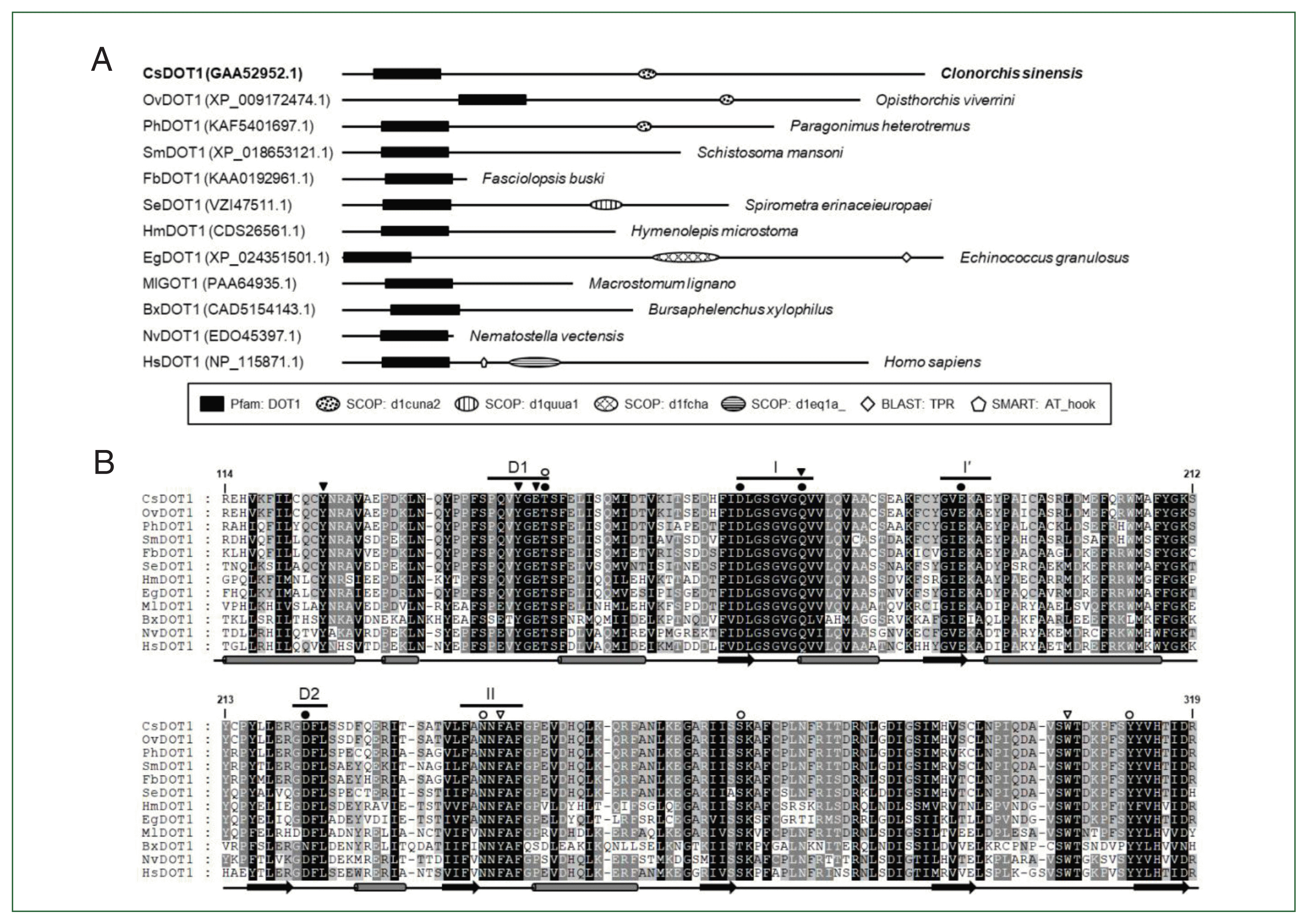 | Fig. 3Structural characterization of Clonorchis sinensis DOT1 (CsDOT1) proteins. (A) The functional domains conserved in CsDOT1 were predicted using the SMART program and compared with those of its orthologs. The domain legends are shown at the bottom. (B) The SET domain sequences of DOT1 homologs shown in panel A were aligned and the degrees of amino acid (aa) conservation were differentially highlighted in the alignment. The secondary structure of the SET domain, which has been determined in human DOT1, is presented at the bottom of each alignment (rod, α-helix; arrow, β-sheet). The segmental motifs constituting the S-adenosyl methionine (SAM)-binding pocket (Motifs I, I′, II, D1, and D2) are shown on the top. Solid and open circles represent aa residues involved in the binding of SAM and Lys79 of histone H3, respectively. Amino acids involved in the formation of a channel leading to the inside of the SAM binding pocket and the regulation of the Lys-binding channel gate are denoted with solid and open arrowheads, respectively. The numerals in the alignment are the positions of corresponding residues in human DOT1 (NP_115871.1). |
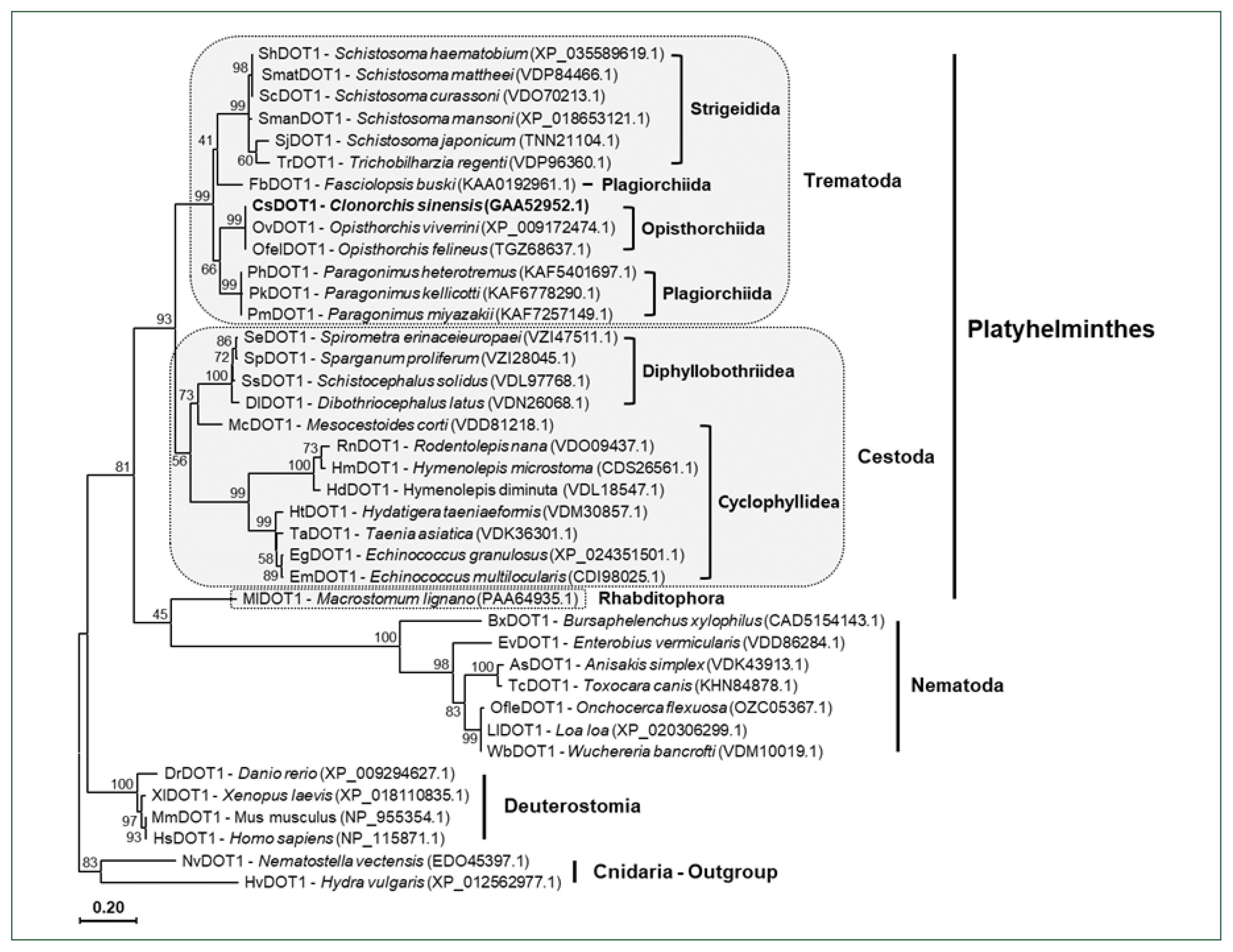 | Fig. 4A phylogenetic tree of platyhelminth and nematode DOT1. The amino acid sequences of the DOT1 domain were isolated from some representative platyhelminth and nematode species including C. sinensis. The sequences were used in the phylogenetic analysis, together with those of deuterostomian and cnidarian orthologs, using the PhyML program. The tree was rooted with the cnidarian DOT1s. Numerals at branching nodes indicate the bootstrap values (1,000 replicas). |
Methylated histones in C. sinensis adults
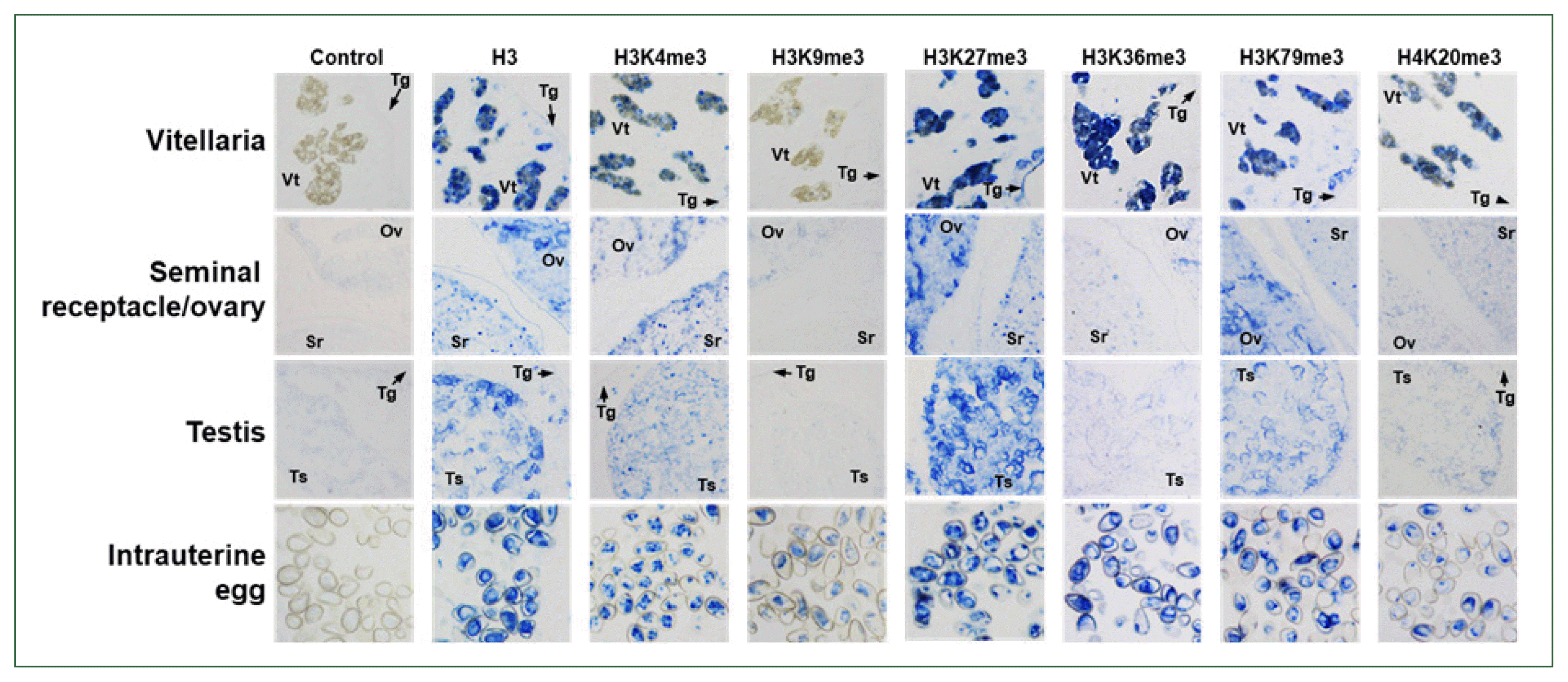 | Fig. 5Immunological detection of methylated histones in Clonorchis sinensis adults. The whole-body sections of C. sinensis adults (42-day-old) were reacted with antibodies specific to histone H3 and its methylated derivatives, as indicated on the top, and the positive reactions were enzymatically stained with the HIGHDEF blue chromogen. Rabbit normal serum was used in the control staining reaction. Ov, ovary; Sr, seminal receptacle; Tg, tegument; Ts, testis; Vt, vitelline follicle. |
Temporal profiling of SET-HKMT gene expressions
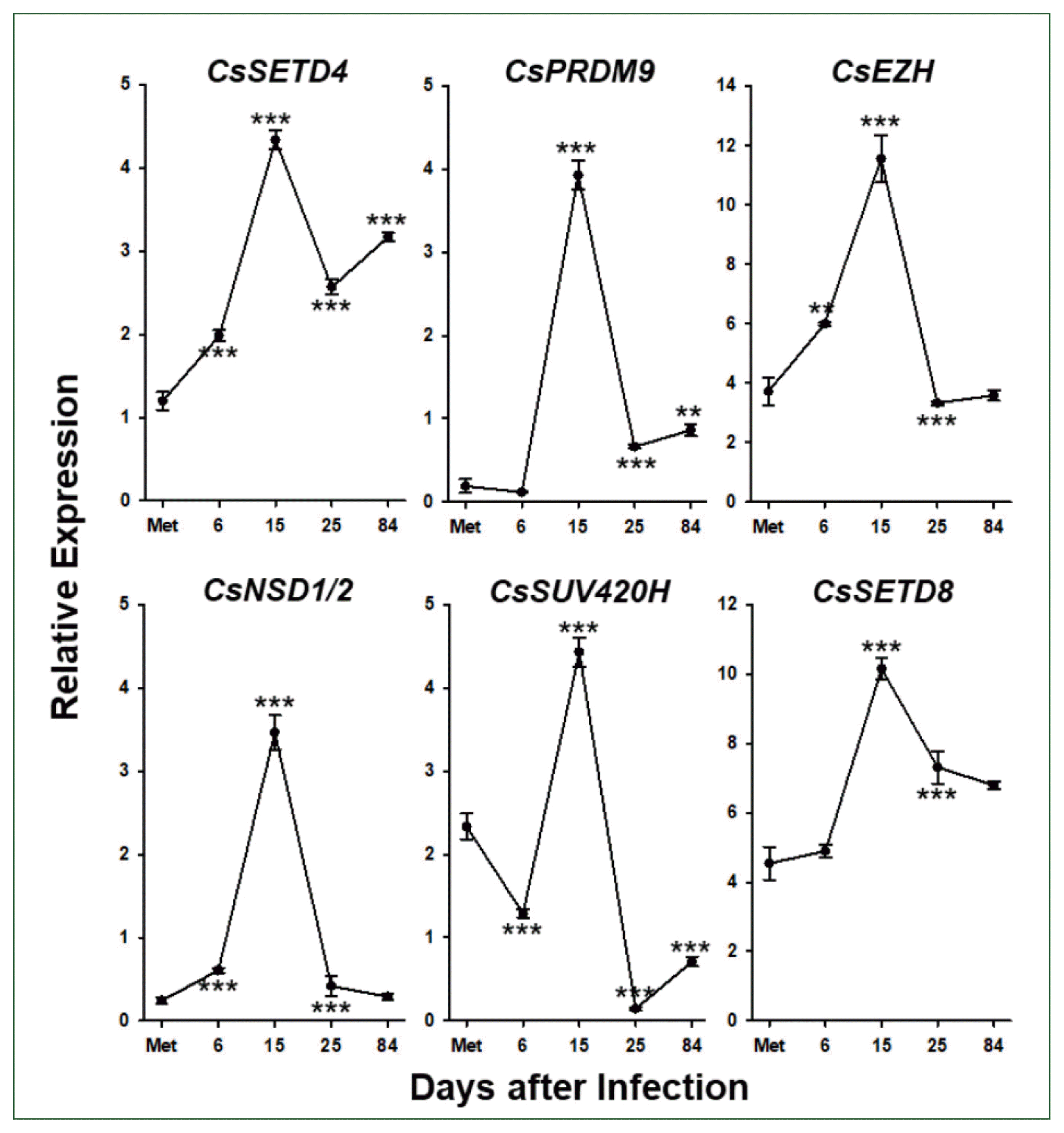 | Fig. 6Temporal expression profiles of Clonorchis sinensis histone lysine methyltransferase (CsHKMT) genes. Expression profiles of the C. sinensis genes were determined using quantitative reverse transcription PCR, in association with the development/maturation of the liver fluke from metacercaria to 84-day-old worm. The calculations are based on independent technical triplicates (n=3, mean±standard deviation). The significance of the expression changes at each stage was statistically estimated by comparing the gene expression levels with those of its direct previous stage using the Student’s t test. **P<0.01, ***P<0.001. |
Effects of HKMT inhibitors on functional gene expression
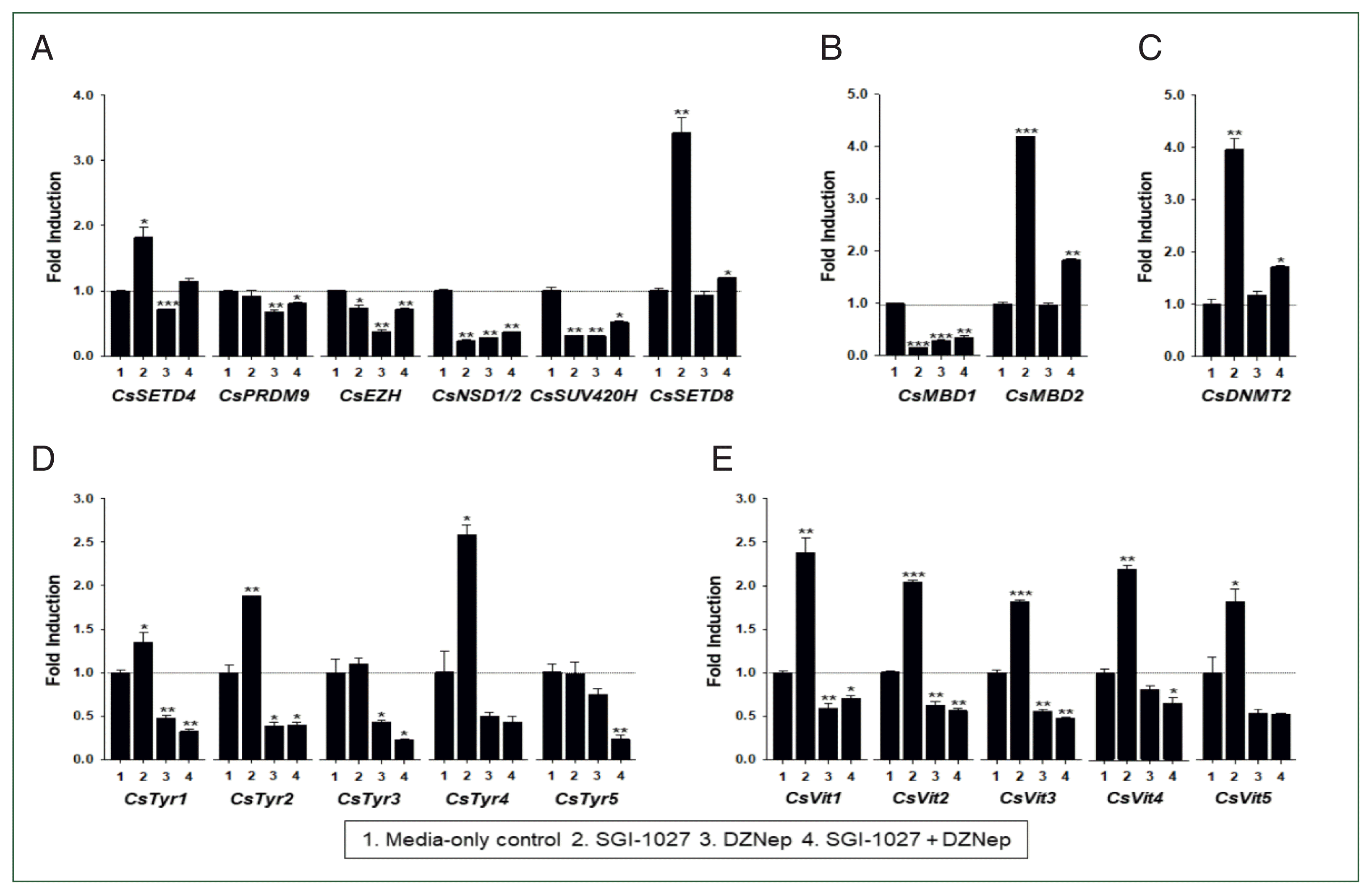 | Fig. 7Effect of methyltransferase inhibitors on functional gene expressions in 140-day-old Clonorchis sinensis adults. Live C. sinensis worms were incubated in the RPMI-1640 medium supplemented with SGI-1027 (5 μm) and/or 3-deazaneplanocin A (DZNep, 0.5 μm) in a 5.0%-CO2 incubator. After a 24 h incubation, total RNAs were extracted from the worms and used in quantitative reverse transcription PCR with primer sets specific to each of the C. sinensis genes. (A) HKMT genes; (B) methyl-CPG; (C) DNMT genes; (D) Tyrosinase genes; (E) eggshell precursor protin genes. Fold increases in the globin gene expression in each of the experimental groups were calculated against the media-only control group (n=3, mean±standard deviation). *P<0.05, **P<0.01, ***P<0.001. |
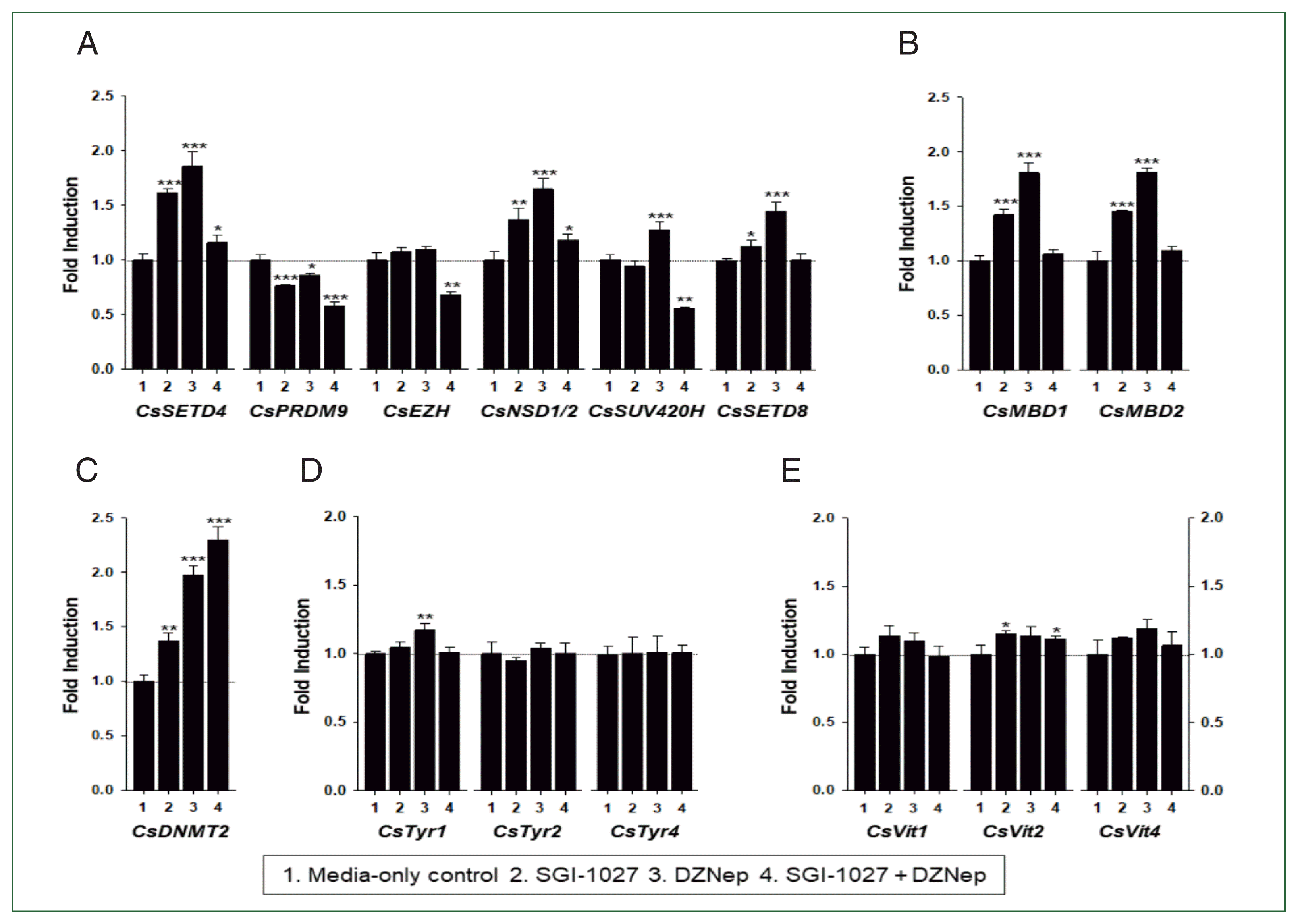 | Fig. 8Effect of methyltransferase inhibitors on functional gene expressions in 42-day-old Clonorchis sinensis adults. Live C. sinensis worms were incubated in the RPMI-1640 medium supplemented with SGI-1027 (5 μm) and/or 3-deazaneplanocin A (DZNep, 0.5 μm) in a 5.0% CO2 incubator. After a 24 h incubation, total RNAs were extracted from the worms and quantitative reverse transcription PCRs were performed using primer sets specific to each of the C. sinensis genes. (A) HKMT genes; (B) methyl-CPG; (C) DNMT genes; (D) Tyrosinase genes; (E) eggshell precursor protin genes. Fold increases in the globin gene expression in each of the experimental groups were calculated against the media-only control group (n=3, mean±standard deviation). *P<0.05, **P<0.01. ***P<0.001. |



