Introduction
Pakistan is a malaria-endemic country that has experienced, due to frequent floods and weak disaster preparedness, a significant rise in malaria cases in recent years. In 2022, the number of suspected malaria cases increased to >3.4 million, compared to 2.6 million estimated cases in 2021. Two
Plasmodium species,
P. falciparum and
P. vivax, have been reported in Pakistan, with the latter predominating (>80%) [
1].
The variant surface antigens (VSAs) of many
Plasmodium spp. are considered the key proteins used by the parasite to escape the host immune system [
2]. These proteins are encoded by multigene families located on telomeric and subtelomeric regions of the parasite,s chromosomes. Similar to the
var gene superfamily in
P. falciparum, the multigene superfamily of variant interspersed repeats (
vir) in
P. vivax encodes proteins used for avoiding the host immune system [
3,
4]. The
vir gene superfamily consists of 346 genes divided into 12 subfamilies A–L [
5]. VSAs are potential targets for vaccine development; however, the lack of an in vitro culture system for
P. vivax and the variant nature of VIRs have presented challenges to their application as a vaccine or diagnostic tool [
6].
Despite the variant nature of VAR proteins, vaccine trials have been partially established [
7]. VIR proteins can protect against sequestration through the use of disrupting rosettes [
8]. Additionally, these proteins induce moderate natural acquisition of antibody and cellular responses in
P. vivax-endemic countries [
9]. Like
var genes,
vir genes are potent candidates for vaccination against vivax malaria [
5]. Since researching the genetic variation of VSAs is important, the genetic diversity of
vir genes in the Republic of Korea (Korea), India, and Myanmar has been previously evaluated [
10–
13]. Here, we analyzed the genetic characteristics of
vir genes in the
P. vivax population of Pakistan and evaluated the population genetics of
vir genes among different Asian populations. Our aim was to obtain in-depth information on the population structure of
P. vivax, which can then be used for the development of vaccines or diagnostic tools.
Discussion
The
VIR proteins are attractive vaccine candidates because they induce a long-lasting antibody response with similar antibody levels in immune sera between singly- and multiply-infected patients without clonal expression [
2]. Moreover, their cell adherence abilities suggest that they play key roles in the pathophysiology of
P. vivax infection [
17,
18]. The genetic diversity patterns in of
VIR proteins in
P. vivax isolates from several countries indicate that
vir genes can be used as genetic markers for the evaluation of genetic polymorphism and transmission hotspots in malaria-endemic regions in the long term [
19]. Therefore, assessing the genetic diversity and population structure in global isolates is important to provide new insights into vaccine development and genetic marker potential.
Consistent with the results of previous studies [
10–
13], we observed different amplification positivity rates among 4
vir genes, which may be due to their location on telomeres and subtelomeres of the
P. vivax genome. Genes in telomeres are very difficult to amplify because of the repetitive nature of telomeric DNA and unusual terminal structure [
20]. Previous analysis of the gene diversity of the 4
vir genes have found that
vir 4 is the most conserved [
11,
14,
15]. However, our analysis of Pakistan isolates found that
vir 4 is the most polymorphic gene with many segregating sites compared to the Sal-1 reference sequence. Furthermore,
vir 21, which is polymorphic in other populations, was conserved among the Pakistan isolates. We attribute our findings to the dynamic molecular evolution of
vir genes due to the sharp increase in malaria cases in Pakistan. This explanation is supported by the results of neutrality analyses of Pakistan
P. vivax isolates, as demonstrated by the wide range of Tajima’s D values in this population (from −2.10836 for
vir 27 to 1.12321 for
vir 4). A positive Tajima’s D value indicates tendencies for an increase in polymorphism, a decrease in population size, and/or balancing selection. A negative Tajima’s D value tendencies for low frequency of polymorphism, increased population size, and/or purifying selection. Additionally, a Tajima’s D value below −2 or above 2 is generally a strong indication that a gene is not evolving neutrally. Genes with a Tajima’s D value below −2 have an excess of rare alleles, indicating positive selection or a selective sweep. However, genes with a Tajima’s D value above 2 have an excess of common alleles suggestive of balancing selection [
21]. In contrast to
vir 4,
vir 27 has a Tajima’s D test value below −2, which indicates unfavorable circumstances for survival and/or in the process of being purged from the gene pool via population expansion, an effect that increases with a lower mutation rate but decreases with a higher recombination rate [
22]. Furthermore, the high diversity of
vir genes, along with a dramatic population expansion, indicates that the
vir genes may play a role in host immune evasion, although more research is needed to explore this possibility.
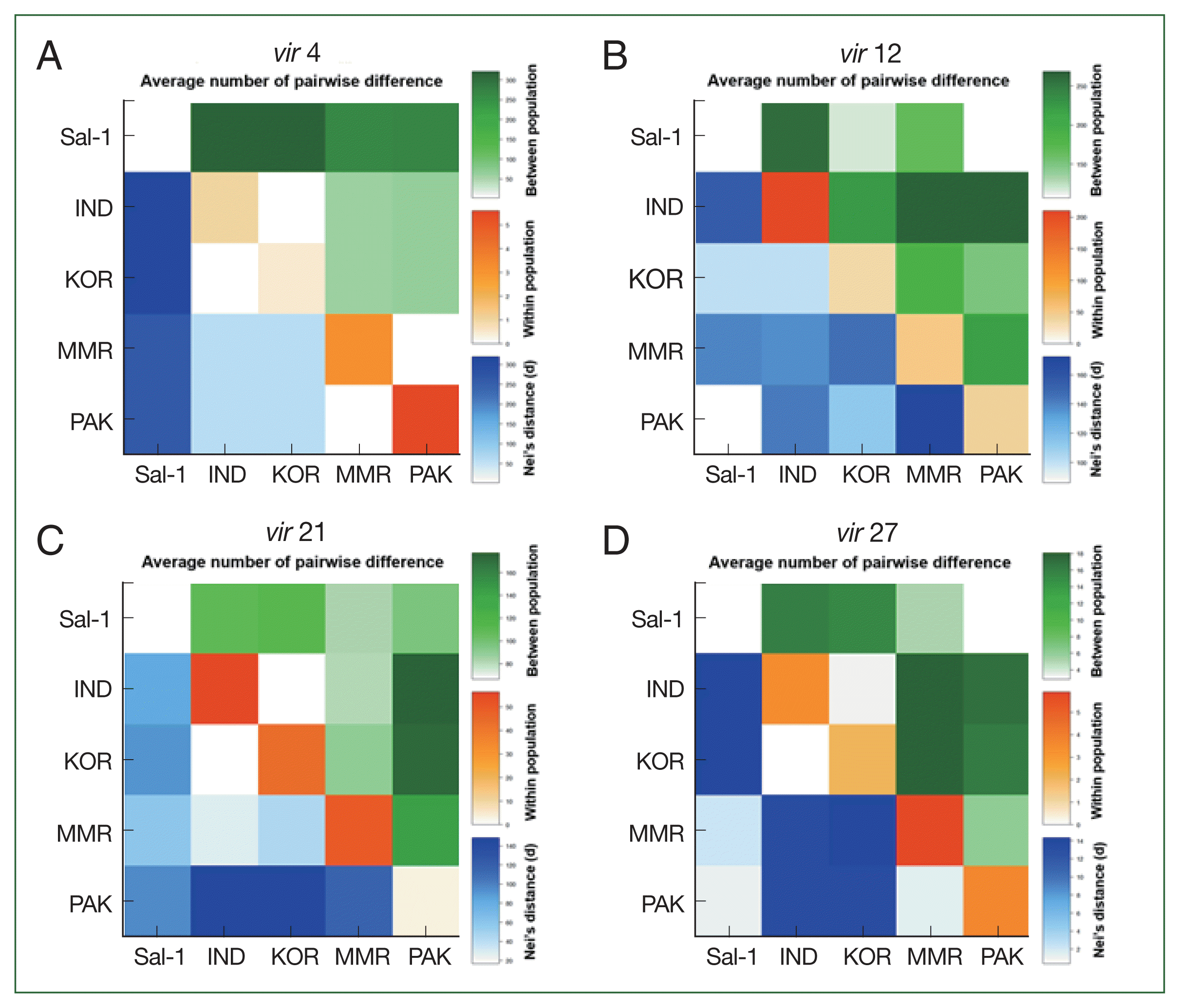
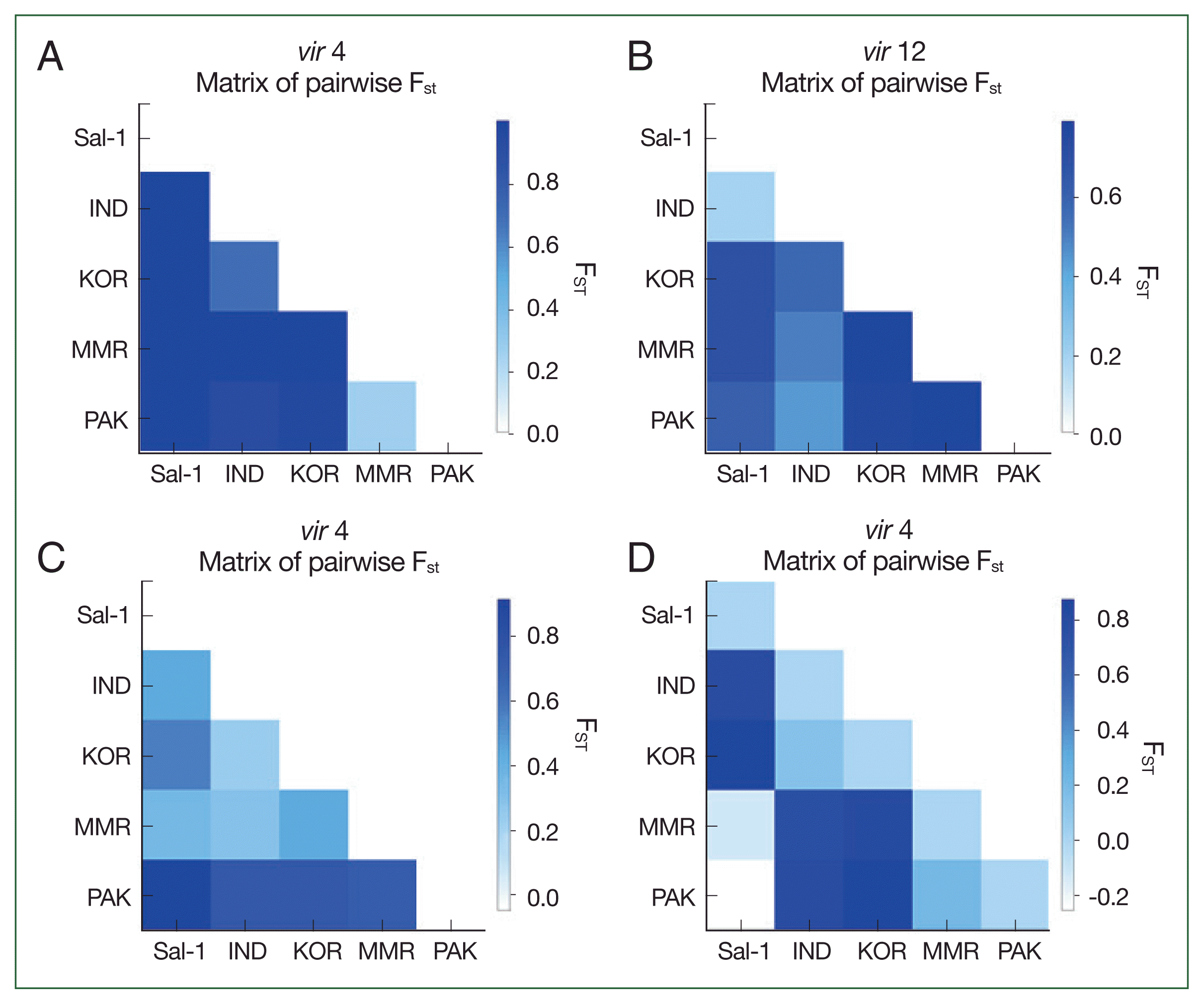
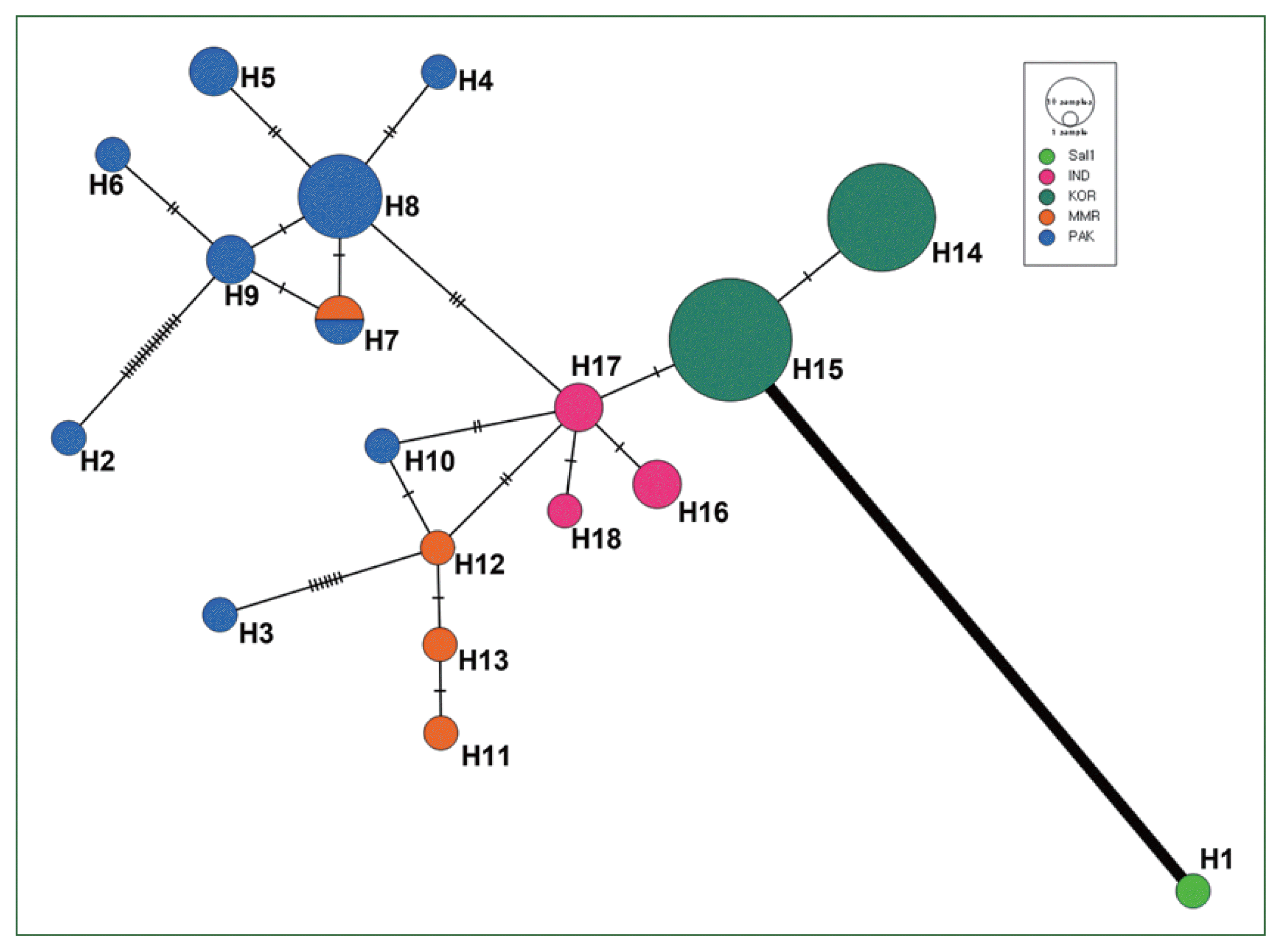
Analyses of pairwise differences and the haplotype network shows that a geographical relationship among global isolates based on the
vir 4 gene, and the Sal-1 reference sequence had a remarkably high pairwise difference and low genetic relationship with other Asian isolates and even haplotypes from each of the Asian countries in the network map (
Fig. 3). This result is supported by the results of AMOVA analysis, which demonstrates high (96.1%) and low (3.9%) percentage variation values among and within populations, respectively, based on
vir 4 characteristics. These results suggest that
vir 4 can be useful as a population marker to evaluate the geographical evolution and genetic structure of
P. vivax. In contrast to
vir 4, we observed the formation of one node consisting of
vir 21 and
vir 27 haplotypes among 5 Asian countries and even Brazil (Sal-1 strain). These close relations are consistent with the relatively low pairwise difference and percentage variation values in AMOVA analysis among populations.
There is currently no useful vaccine for
P. vivax malaria; however,
vir genes are vaccine candidates [
23]. Despite this possibility, there are only limited studies on the genetic diversity and population relationships of
vir genes at the global level. Longitudinal genetic diversity studies of
vir genes in
P. vivax populations from diverse countries are necessary to further understand
vir gene function and its application to malaria control, e.g., as vaccines.
In the present study, we investigated the genetic diversity and relationships between populations of P. vivax from 5 countries (Pakistan, Myanmar, India, Korea, and Brazil) based on analysis of 4 vir genes. Among the 4 genes, vir 4 showed remarkable genetic differentiation among populations, although its characteristics were conserved within populations, suggesting that vir 4 can be a potent marker for understanding P. vivax population structure. In contrast, vir 21 and vir 27 are characterized by a low pairwise difference and percentage variation among populations; therefore, these can be considered vaccine candidates. A better understanding of the function of vir genes and their application to malaria control, e.g., as vaccines, requires longitudinal genetic diversity studies of vir genes in samples obtained from more diverse countries.