Abstract
Advancements in the field of proteomics have provided great opportunities for the development of diagnostic and therapeutic tools against human diseases. In this study, we analyzed haptoglobin and amyloid A protein levels of vivax malaria patients with combinations of depletion of the abundant plasma proteins, 2-dimensional gel electrophoresis (2-DE), image analysis, and mass spectrometry in the plasma between normal healthy donors and vivax malaria patients. The results showed that the expression level of haptoglobin had become significantly lower or undetectable in the plasma of vivax malaria patients due to proteolytic cleavage when compared to healthy donors on 2-DE gels. Meanwhile, serum amyloid A protein was significantly increased in vivax malaria patient's plasma with high statistical values. These 2 proteins are common acute phase reactants and further large scale evaluation with a larger number of patient's will be necessary to establish the possible clinical meaning of the existential changes of these proteins in vivax malaria patients. However, our proteomic analysis suggests the feasible values of some plasma proteins, such as haptoglobin and serum amyloid A, as associating factor candidates for vivax malaria.
-
Key words: Plasmodium vivax, vivax malaria, proteomics, associating factor candidate, haptoglobin, serum amyloid A
INTRODUCTION
Malaria, which is caused by the apicomplexan parasite
Plasmodium spp., is one of the most important parasitic diseases globally, with approximately 200-300 million clinical cases and 1-3 million deaths each year worldwide [
1,
2]. Of the 4 human infectious species,
Plasmodium vivax and
P. falciparum are the major parasites, causing up to 90% of malaria cases [
3].
P. vivax is the most widely distributed human malaria parasite, which is prevalent in South America, Asia, and Oceania. The parasite causes 80-300 million cases every year, with clinical manifestations ranging from mild to chronic infections that in some cases lead to severe complications or death.
To control malaria, accurate and prompt diagnostic methods are urgently needed. In general, two classical methods are used to diagnose malaria; clinical symptoms and microscopic observation of a blood smear. However, microscopic examination requires technically-skilled personnel and clinical symptoms are not specific [
4]. PCR is the most sensitive diagnostic tool; however, it suffers from the important shortcoming of being difficult to use in the field [
5].
In seeking an alternative diagnostic approach, protein biomarkers are a candidate worthy of exploration. The use of protein biomarkers for the early diagnosis or monitoring of treatment for chronic diseases, including cancer, autoimmune diseases, and infectious diseases is now a central research interest in clinical proteomics, particularly for the identification of proteins that may respond to changes in disease states or drug treatments in easy-to-access and clinically meaningful biofluids, such as human plasma. Plasma proteins often represent the most comprehensive physiological state of the body, as they originate from different tissues and organs through diffusion or secretion upon infection, disease, or tissue injury [
6]. Accordingly, numerous studies have done to identify diagnostic plasma protein markers for cancer, metabolic diseases, and other symptomatic diseases.
The proteomic approach is a powerful tool that can provide protein expression profiles, which may useful to predict clinical events, therapeutic response, or to probe underlying mechanisms of diseases, such as autoimmune disorders, cardiovascular diseases, and cancers [
7-
10]. The urgent need for the proteomic-based discovery of novel disease markers and/or evaluating factors associated with specific diseases together with the development of high-throughput techniques that provide highly sensitive analyses of the protein content in cells, tissues, and organisms, as well as of different body fluids, such as plasma and urine, has opened a completely new chapter in biomarker discovery. Biomedical proteome research aimed at biomarker discovery is mainly based on expression proteomics, which analyzes the quantity of certain proteins in different conditions. Thus, proteomic studies that aid characterization of proteins and selection of optimal proteomics technologies are likely to be key factors in propelling the discovery of novel biomarkers.
With this goal, in this study, we analyzed plasma proteins of patients with vivax malaria and healthy subjects to discover biomarkers for vivax malaria and in-depth understanding of pathophysiological mechanisms of the disease by using combinations of depletion of the abundant plasma proteins, 2-dimensional gel electrophoresis (2-DE), image analysis, and mass spectrometry (MS). Hitherto, there has been no report concerning the proteomic analysis of plasma proteins from patients infected with vivax malaria. The results possibly indicate that some plasma proteins, in particular haptoglobin and serum amyloid A, could be developed as possible biomarkers or associating factor candidates for vivax malaria, pending clarification of their clinical meaning.
MATERIALS AND METHODS
Chemicals
Immobilized pH gradient (IPG) strips of pH 3-10 (nonlinear), 4-7, 4.5-5.5, and 5.5-6.7 (Immobiline DryStrip, 0.5 × 3 × 180 mm) were purchased from GE HealthCare Bioscience (Uppsala, Sweden). Sodium dodecyl sulfate (SDS), acrylamide, bis-acrylamide, tetramethylethylenediamine, Tris, glycine, glycerol, and formaldehyde were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Dithiothreitol (DTT), urea, CHAPS, Ready Sol IEF 40% solution™, and IPG buffers were also from GE HealthCare Bioscience. Silver nitrate was obtained from Fluka Chemie AG (Steinheim, Switzerland), and sequencing grade trypsin was from Roche Diagnostics (Mannheim, Germany). Other reagents for 2-DE and MS were purchased from Sigma-Aldrich or Merck (Whitehouse Station, New Jersey, USA).
Plasma collection
Blood samples of patients with vivax malaria were kindly provided from the National Institute of Health, Korea. The study population consisted of outpatients with acute febrile syndrome visiting the community health centers of Northern Gyeonggi-do (Province). Thin and thick blood smears were prepared for microscopic diagnosis from blood collected from individuals' fingertips. A local health unit employee read the Giemsa-stained blood smears and the observations confirmed by the following microscopic examination. For preparation of plasma samples, whole blood was collected by venipuncture into BD Vacutainer serum separation tube (BD Biosciences, San Jose, California, USA), centrifuged at 1,300 g for 10 min at room temperature (RT), after which plasma was pooled, aliquoted, and stored in a liquid nitrogen tank or at -80℃. Total protein concentrations of plasma samples were estimated using a bicinchoninic acid-based protein assay system (Pierce Chemical, Rockford, Illinois, USA). Authorization for the use of all of plasma for protocol approval and ethical clearance was approved by the Institutional Review Board of Inha University School of Medicine.
Depletion of albumin and IgG from plasma
Human plasma samples of 35 µl were individually diluted 10-fold with ProteoExtract™ albumin/IgG binding buffer (Cal-Biochem, San Diego, California, USA), filtered through a 0.22 µm microcentrifuge filter tube, and passed through a resin bed by gravity-flow according to the manufacturer's instructions. The column was washed twice with the ProteoExtract™ albumin/IgG binding buffer and the second wash was collected as flow-through and concentrated using a 5 K molecular weight cut-off spin concentrator. The concentrated pool was either used immediately or aliquoted and stored at -80℃ for future use.
Depletion of six high abundant proteins in human plasma
To remove six major plasma proteins (albumin, transferrin, IgG, IgA, haptoglobin, and anti-trypsin) in human plasma, the multiple affinity removal column system (MARS®; 4.6 × 50 mm; Agilent Technologies, Wilmington, Delaware, USA) was routinely used. Briefly, 20 µl of human plasma was diluted with 80 µl Buffer A (equilibration buffer), filtered through a 0.22 µm microcentrifuge filter tube, and injected onto the antibody column at a flow rate of 0.25 ml/min. The flow-through fractions containing unbound proteins from the injections were collected, pooled, and concentrated using a 5 K molecular weight cutoff spin concentrator. The concentrated pool was either used immediately or aliquoted and stored at -80℃ for future use.
Depletion of 20 high abundant proteins in human plasma
For removal of the top 20 most abundant proteins in human plasma, plasma was applied to the ProteoPrep 20® plasma immunodepletion kit (Sigma-Aldrich). This kit is specifically designed to remove 20 highly abundant proteins, namely albumin, transferrin, α1-acid glycoprotein, complement C1q, IgG, fibrinogen, ceruloplasmin, complement C3, IgA, α2-macroglobulin, apolipoprotein A-I, complement C4, IgM, α1-antitrypsin, apolipoprotein A-II, plasminogen, IgD, haptoglobin, apolipoprotein B, and prealbumin. To accomplish protein depletion, 8 µl of human plasma was diluted to 100 µl with equilibration buffer, filtered through a 0.22 µm microcentrifuge filter tube, and loaded onto the antibody resin in a microcentrifuge spin column following the manufacturer's instructions. The sample was either passed into the column at a slow rate (1.5 min, 100 g, RT) or incubated at 4℃ for 15 min. In both cases, the column was washed twice with equilibration buffer and centrifuged (2.5 min, 100 g) to collect the total unbound protein plus the wash fractions. The flow-through fraction containing unbound proteins was concentrated with a 5 K molecular weight cutoff spin concentrator for immediate analysis or aliquoted and stored at -80℃ for future use.
Two-dimensional gel electrophoresis (2-DE)
An appropriate amount (1-3 µl) of whole or depleted plasma was rehydrated, focused, and then applied to the second dimension essentially as described previously [
11,
12]. Briefly, plasma samples were thawed and diluted into an isoelectric focusing (IEF) buffer containing 9 M urea, 4% CHAPS, 0.1 M DTT, 0.8% ampholyte buffer and 1X protease inhibitor cocktail™ (Roche) to yield the desired protein amount in a volume that could be absorbed by IPG strips [pH gradients of 3-10 (nonlinear) (total 80 kVh), 4-7 (92 kVh), 4.5-5.5 (120 kVh), or 5.5-6.7 (120 kVh)], typically using 350 µl for an 18 cm IPG strip, followed by electrophoresis in the second dimension performed by 9-12% gradient SDS-PAGE. Gels were silver-stained according to Yan et al. [
11], which is compatible with trypsin digestion and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). Briefly, each gel was fixed with methanol/acetic acid/water (40 : 10 : 50) for 30 min, followed by sensitizing in 30% methanol, 5% sodium thiosulfate, and 6.8% (w/v) sodium acetate for 30 min. This was followed by 3 times 5 min washes in deionized water. Proteins were stained in a solution of 2.5% silver nitrate for 20 min, and the gel was washed twice in deionized water for 1 min each time. Subsequently, the gel was developed with 2.5% (w/v) sodium carbonate and 0.04% formaldehyde. When the desired intensity was attained, the developer was discarded and the reaction was stopped with 1.46% (w/v) EDTA for 10 min. The gel was washed with deionized water several times and stored in a sealed plastic bag at 4℃. Protein patterns in the gel were recorded as digitalized images using a GS-800 Calibrated Imaging Densitometer® high-resolution scanner (Bio-Rad, Hercules, California, USA). Scanned images were analyzed with the Melanie IV® 2-DE program, (Geneva Bioinformatics, Geneva, Switzerland).
Following the selection of the protein spots of interest, the spots were manually excised from the stained 2-DE gels and subjected to in-gel digestion [
11,
13,
14]. In-gel digestion of protein spots on silver-stained gels was performed as previously described [
12] with minor modification. In brief, protein spot gel pieces for MS analysis were excised and put into microcentrifuge tubes. Prior to in-gel digestion, each excised spots were destained in a 1 : 1 mixture of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate with gentle shaking for 10 min. The particles were washed twice with deionized water with gentle shaking for 15 min. The destained gels were washed with 50% acetonitrile (ACN) and 25 mM ammonium bicarbonate, pH 7.8, with gentle shaking for 10 min. The liquid was removed and ACN (50 µl) was added and incubated for 5 min. All liquid was removed and the gel particles were dried in a vacuum centrifugal concentrator for 5 min. The gel particles were individually incubated on ice in 10 µl of 0.02 µg/ml sequencing grade trypsin solution. After 45 min, the supernatant was discarded and replaced with 20 mM ammonium carbonate. After 12 h of digestion at 37℃, 10 µl of 0.5% (v/v) trifluoroacetic acid (TFA) in 50% ACN was added and the solutions were sonicated twice in an ultrasonic water bath. The extraction was performed with 0.1% formic acid in 2% ACN for MALDI-TOF MS.
Protein spots of interest were subjected to MALDI-TOF MS. Mass analysis was performed on a PerSeptive Biosystem Voyager-DE STR™, MALDI-TOF MS (Applied Biosystems, Foster City, California, USA) in reflector mode for positive ion detection. The spectrometer was run with the following settings: accelerating voltage, 20 kV; grid voltage, 65%; and a DELAY of 100 NS. Each tryptic peptide extract (0.5 µl) was dispensed onto a MALDI sample plate along with 0.5 µl of matrix solution consisting of 10 mg/m α-cyano-4-hydroxycinnamic acid, 0.1% TFA, and 50% ACN. External peptide calibrants, angiotensin I (monoisotopic mass, 1296.6853), rennin substrate (1758.9331), and adrenocorticotropic hormone (2465.1989) were used for mass calibration. Spectra were internally calibrated using autolytic fragments from trypsin. Proteins were identified by peptide mass fingerprinting (PMF) with the search engine programs ProFound and Mascot. All mass searches were performed using a mass window between 0 and 100 kDa.
Validation of the target proteins
Validation of some differentially-expressed proteins was assessed by Western blot analyses with the commercially available specific antibody against the target protein. Thirty micrograms of whole plasma protein was separated by 10% SDS-PAGE. Proteins were electrophoretically transferred onto a PROTEAN® nitrocellulose membrane (Whatman GmbH, Dassel, Germany) using Semi-Dry Transfer Blotter (GE HealthCare Bioscience), blocked with 5% nonfat dry milk in Tris-buffered saline (TBS), and subsequently identified using a specific antibody against target protein followed by horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad). The primary polyclonal antibody against serum amyloid A protein (sc-20651) was purchased from Santa Cruz Biotechnologies (Santa Cruz, California, USA). The immunoreactive proteins on the membrane were detected by chemilluminescence using the West-Save™ substrate (AbFrontier, Seoul, Korea) onto X-ray film (Agfa-Gebaert N.V., Mortsel, Belgium).
RESULTS
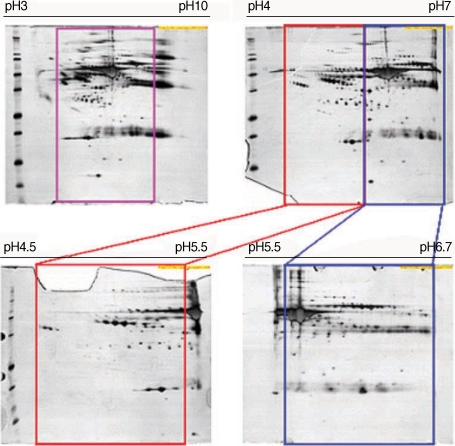
To define optimal condition for 2-DE, 2-DE analysis was carried out using plasma from normal healthy donors in different experimental conditions. These results showed that isoelectric focusing using the pH range of 3-10 (nonlinear) was optimal (
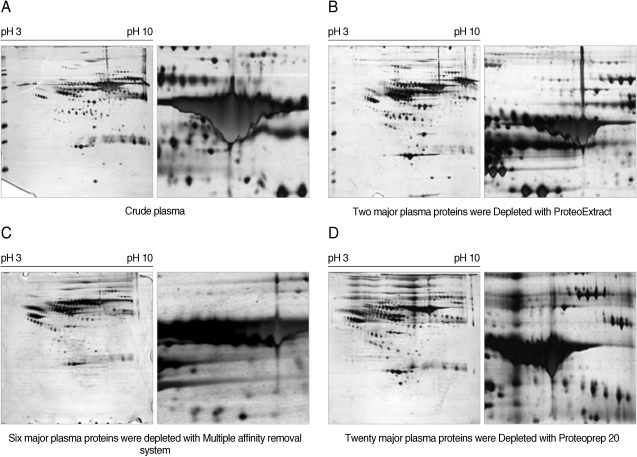
Fig. 1). The effectiveness of major protein depletion using several different methods is shown in
Fig. 2. Disease-specific biomarkers hold diagnostic promise in both human and veterinary medicine. However, the large dynamic range of proteins in serum makes the analysis very challenging because high-abundant proteins tend to mask those of lower abundance. A prefractionation step, such as depletion of a few high-abundant proteins before protein profiling, can assist in the discovery and detection of less abundant proteins that may prove to be informative biomarkers. Different commercially available protein-partitioning products were tested for their ability to lower the detection limit of proteins in 2-D gels (
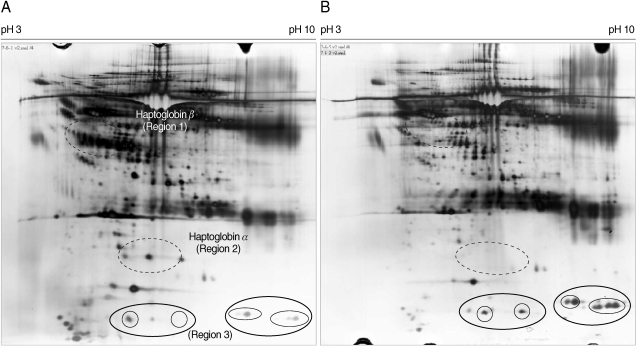
Fig. 2). Next, differential changes in plasma proteins between malaria patients and healthy donors were analyzed using 2-DE. Four plasma samples from two normal donors and two vivax malaria patients were separated by 2-DE analysis using the pH 3-10 range and the resulting patterns were compared. Several protein spots in the 2-DE gels were significantly and consistently different in the plasma from the vivax malaria patients when compared with plasma from the normal healthy donors (
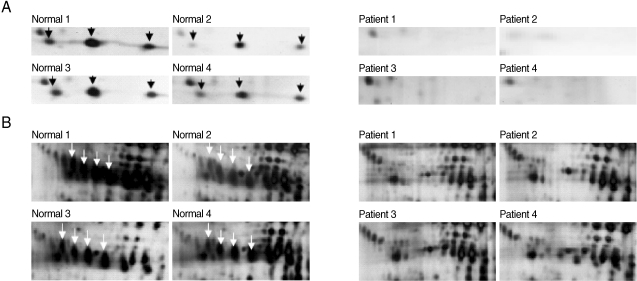
Fig. 3). The most significant differences were identified in regions 1, 2, and 3 (
Figs. 3,
4). A group of proteins in these regions were identified as haptoglobin when analyzed with MALDI-TOF MS followed by the ProFound and Mascot search engine programs. Four protein spots in region 1, which were detected as a series of protein spots with different pI values, were identified as haptoglobin β-chain, whereas three protein spots in region 2 were identified as truncated α-chain of haptoglobin (
Figs. 3,
4). Haptoglobin levels in the plasma from vivax malaria patients were significantly lower or had become undetectable, when compared to those of the normal healthy donors on the 2-DE gels (
Figs. 3,
4). The truncated α-chain of haptoglobin was also appeared in region 3 in vivax malaria plasma with increased lower molecular mass protein spots, which may correspond to the fragments probably produced by proteolysis (
Figs. 3,
4).
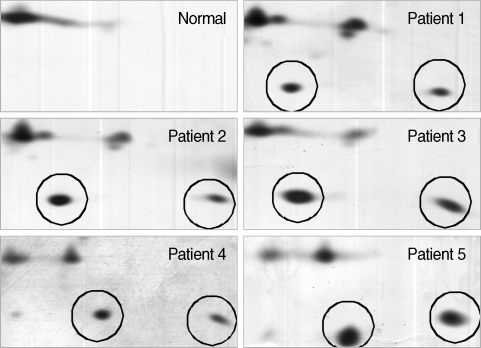
After the removal of six abundant proteins, the plasma proteomes from the vivax malaria patients and the healthy donors were compared by 2-DE. One protein showing a tendency to increase in vivax malaria plasma was serum amyloid A (SAA). Two protein spots were conclusively identified as SAA on the gels resolving the plasma proteome from one normal healthy donor and the plasma of five vivax malaria patients and they were significantly increased in volume and density (
Fig. 5).
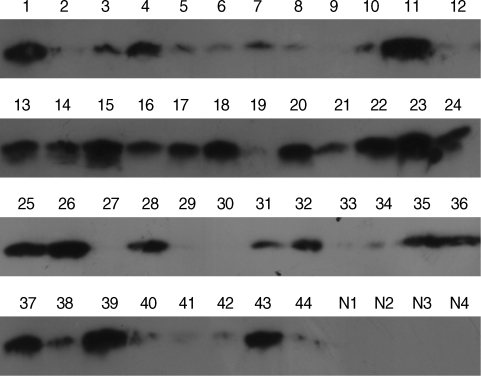
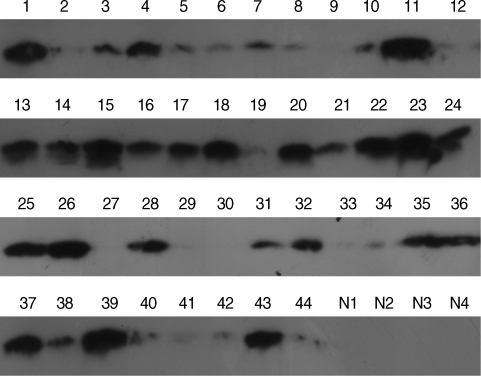
As anticipated, these results were consistent with those from the original 5 plasma samples analyzed by 2-DE and showed that SAA expression in the vivax malaria patient plasma was increased (
Fig. 6). A total of 40 of 44 samples (90.9%) displayed a positive signal at the position corresponding to a molecular weight of approximately 15 kDa, the molecular weight of SAA, indicating that SAA was significantly increased of its protein expression level in the plasma from vivax malaria patients by the Western blot analysis with the commercially available specific antibody against SAA protein. Meanwhile, no detectable signal was identified in the plasma from 4 healthy donors (
Fig. 6).
DISCUSSION
Over the past few years, proteomics has proved to be an extremely promising method of protein analysis. Recent proteomic studies of body fluid, especially blood, has been achieved and is expected to lead to the development of new non-invasive tests and procedures, along with advantage of high efficiency of patient care and of early detection for non-invasive diagnostics in many diseases. Adjustment of proteomic technologies will enable its application at the patient bedside, which would direct medical practice toward personalized medicine.
One of the main goals of this study was to analyze the plasma proteins of human patients with vivax malaria by proteomic approaches, 2-DE, and mass spectrometry to discover biomarkers or associating factor candidates for vivax malaria. To achieve this goal, establishing the optimal experimental condition for 2-DE analysis of human plasma was essential. Efficient separation of clinically relevant proteins that may define disease stages and/or predict disease development is the most critical aspect of biomarker discovery. Because biomarkers are usually low in abundance, depletion of the highly abundant proteins in human serum or plasma, e.g. albumin, that account of the large proportion (approximately 55%) of plasma proteins, can provide an efficient means of separating biomarkers from more abundant proteins. In this study, 2-DE analysis of human plasma was carried out after depleting 2, 6, or 20 of the most abundant proteins in plasma to define the optimal study condition. Differentially-expressed spots were identified by MALDI-TOF MS. Through the removal of the abundant plasma proteins and ultra membrane centrifugation, well-resolved and reproducible 2-DE profiles were obtained.
Many proteins in plasma display complex combinations of post-translational modifications (particularly involving glycosylation) that can be discriminated by 2-DE [
15]. In the haptoglobin chain, three heterogeneities superimpose to create a complex but interpretable pattern of at least 43 resolved spots [
16]. As the results, we identified two proteins, which showed significant difference in their expression level between the plasma from vivax malaria patients and normal healthy donors. Haptoglobin levels in the plasma from vivax malaria patients were significantly lower or had become undetectable compare to those of the normal healthy donors. Meanwhile, SAA was significantly increased in the plasma from vivax malaria patients.
Haptoglobin is an acute-phase response serum protein that plays an important inhibitory role in inflammation [
17]. Intravascular hemolysis is a physiological phenomenon as well as a severe pathological complication when accelerated in various autoimmune, infectious, and inherited disorders. Haptoglobin released into plasma is captured by the acute phase protein haptoglobin, which is depleted from plasma during elevated hemolysis [
18]. When hemoglobin (Hb) is outside of the red blood cell, free haptoglobin binds to Hb and the haptoglobin-Hb complex is cleared by macrophages and monocytes. In malaria infection, red blood cell rupture will release abundant amounts of Hb. Free haptoglobin would subsequently disappear as the haptoglobin-Hb complexes formed. Hypohaptoglobin has been reported in falcipalum malaria infection [
16,
19]. Human SAA is a 12-14 kDa protein whose levels can increase up to 1,000-fold in the serum 24-36 hr after infection or injury, decline after 4-5 days, and then return to baseline after 10-14 days [
20]. The synthesis of acute-phase protein SAA is largely regulated by inflammation-associated cytokine-peptide hormone signals and a high concentration of circulating SAA may represent an ideal marker for inflammatory disease tissue injury, infection, and inflammation [
21]. Human SAA proteins are a group of apolipoproteins found predominantly in the high-density lipoprotein (HDL) fraction of plasma; they are a precursor to amyloid A protein, which is the major constituent of the fibril deposits of reactive amyloidosis. Whereas acute inflammation has mainly beneficial effects, clinical and epidemiological studies have suggested a strong association between chronic infection, chronic inflammation, and cancer [
22]. Basically, SAA is found at very low levels in the sera of healthy donors [
23]. Thus, high SAA levels may be apparently due to occult inflammatory and neoplastic diseases. Previously, Gillespie et al. [
24] reported in the study with 17 adult patients with acute falciparum malaria that concentrations of acute phase reactants, C-reactive protein and SAA, were increased. The authors wanted to test whether measurement of acute phase reactants as an indirect measure of cytokine concentration would prove valuable in assessing the severity of falciparum malaria. Next, to verify the identities of proteins deduced from the results of MALDI-TOF MS, the expression levels of human SAA were analyzed by Western blot using four additional healthy donors and plasma from 44 vivax malaria patients. Thirty micrograms of whole plasma protein were analyzed by Western blotting using a commercially available anti-SAA antibody and the intensity of the chemilluminescence response bands was evaluated.
In conclusion, in the present study, we mapped differently expressed proteins in plasma of vivax malaria patients using proteomic techniques, which allowed the identification of changes of two proteins. Our results demonstrated that haptoglobin was significantly lower (or disappeared) in plasma of vivax malaria patients when compared to those of the normal healthy donors, but SAA expression was significantly increased in vivax malaria patients. Although the changes of the two proteins are not likely to be specific for vivax malaria patients, our study suggested the methological advances for proteomic approach of plasma proteins in vivax malaria patients. Further efforts to identify specific plasma proteins caused by vivax malaria should be performed.
ACKNOWLEDGEMENTS
This research was supported by grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Basic Science Research Program, 313-2008-2-E00184) to TSK, from the National Core Research Center for Nanomedical Technology through Yonsei University (R15-2004-024-02002-0) to KJL, and from the Korean Research WCU grant (R31-2008-000-10086-0) and the National Institute of Health, Ministry of Health and Welfare, Korea (2008-E00272-00) to YYB.
References
- 1. Sachs J, Malaney P. The economic and social burden of malaria. Nature 2002;415:680-685.
- 2. Hemingway J, Bates I. Malaria: past problems and future prospects. EMBO Reports 2003;4:S29-S31.
- 3. Galinski MR, Barnwell JW. Plasmodium vivax merozoite invasion of reticulocytes and considera-tions for malaria vaccine development. Parasitol Today 1996;12:20-29.
- 4. Richardson DC, Ciach M, Zhong KJY, Crandall I, Kain KC. Evaluation of the Markromed dipstick assay versus PCR for diagnosis of Plasmodium falciparum malaria in returned travelers. J Clin Microbiol 2002;40:4528-4530.
- 5. Snounou G, Viriyakosol S, Zhu XP, Jerra W, Pinheiro L, Rosaria VE, Thaithong V, Brown KN. High sensitivity of detection of human malaria parasites by the use of nest polymerase chain reaction. Mol Biochem Parasitol 1993;61:315-320.
- 6. Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res 2005;4:1073-1085.
- 7. Mateos-Caceres PJ, Garcia-Mendez A, Lopez Farre A, Macaya C, Nunez A, Gomez J, Alonso-Orgaz S, Carrasco C, Burgos ME, de Andres R, Granizo JJ, Farre J, Rico LA. Proteomic analysis of plasma from patients during an acute coronary syndrome. J Am Coll Cardiol 2004;44:1578-1583.
- 8. Hershko AY, Naparstek Y. Autoimmunity in the era of genomics and proteomics. Autoimmun Rev 2006;5:230-233.
- 9. Albitar M, Potts SJ, Giles FJ, O'Brien S, Keating M, Thomas D, Clarke C, Jilani I, Aguilar C, Estey E, Kantarjian H. Proteomic-based prediction of clinical behavior in adult acute lymphoblastic leukemia. Cancer 2006;106:1587-1594.
- 10. Hudelist G, Singer CF, Pischinger KI, Kaserer K, Manavi M, Kubista E, Czerwenka KF. Proteomic analysis in human breast cancer: identification of a characteristic protein expression profile of malignant breast epithelium. Proteomics 2006;6:1989-2002.
- 11. Yan JX, Wait R, Berkelman T, Harry RA. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. Electrophoresis 2000;21:3666-3672.
- 12. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 1996;68:850-858.
- 13. Park JW, Kim S, Bahk YY. A proteomic approach for dissecting H-Ras signaling networks in NIH/3T3 mouse embryonic fibroblast cells. Proteomics 2006;6:2433-2443.
- 14. Kim S, Lee YZ, Kim YS, Bahk YY. A proteomic approach for protein-profiling the oncogenic ras induced transformation (H-, K-, and N-Ras) in NIH/3T3 mouse embryonic fibroblasts. Proteomics 2008;8:3082-3093.
- 15. Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Identification of the haemoglobin scavenger receptor. Nature 2001;409:198-201.
- 16. McQuire W, D'Alessandro U, Olaleye BO, Thomson MC, Langerock P, Greenwood BM, Kwiakowski D. Creactive protein and haptoglobin in the evaluation of a community-based malaria control programme. Trans R Soc Trop Med Hyg 1996;90:10-14.
- 17. Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 2002;1:845-867.
- 18. Anderson NL, Anderson NG. Microheterogeneity of serum transferrin, haptoglobin and 2 HS glycoprotein examined by high resolution two-dimensional electrophoresis. Biochem Biophys Res Commun 1979;8:258-265.
- 19. Baumann H, Gauldie J. The acute phase response. Immunol Today 1994;15:74-80.
- 20. Gabay C, Kushner I. Mechanisms of disease: acute-phase proteins and other systemic responses to inflammation. N Eng J Med 1999;340:448-454.
- 21. Malle E, Sodin-Semrl S, Kovacevic A. Serum amyloid A: An acutephase protein involved in tumor pathogenesis. Cell Mol Life Sci 2009;66:9-26.
- 22. Liu DH, Wang XM, Zhang LJ, Dai SW, Liu LY, Liu JF, Wu SS, Yang SY, Fu S, Xiao XY, He DC. Serum amyloid A protein: a potential biomarker correlated with clinical stage of lung cancer. Biomed Environ Sci 2007;20:33-40.
- 23. d'Eril GM, Anesi A, Maggiore M, Leoni V. Biological variation of serum amyloid A in healthy subjects. Clin Chem 2001;47:1498-1499.
- 24. Gillespie SH, Dow C, Raynes JG, Behrens RH, Chiodini PL, Mc-Adam KPWJ. Measurement of acute phase proteins for assessing severity of Plasmodium falciparum. J Clin Pathol 1991;44:228-231.
Fig. 1Comparison of 2-DE maps of whole plasma from normal healthy donors in different experimental conditions. Plasma samples obtained from healthy normal individuals were analyzed by 2-DE as described in Materials and Methods. For these maps, 100 µg (pH range of 3-10), 150 µg (4-7), 200 µg (4.5-5.5) and 200 µg (5.5-6.7) of whole plasma proteins were applied by in-gel rehydration and to different IPG strips, pH ranges of 3-10 (nonlinear), 4-7, 4.5-5.5, and 5.5-6.7. For separation in the second dimension, 9-12% gradient SDS-PAGE was used. Three or more independent runs of each plasma sample were analyzed for each condition. The gels were visualized by silver nitrate staining.
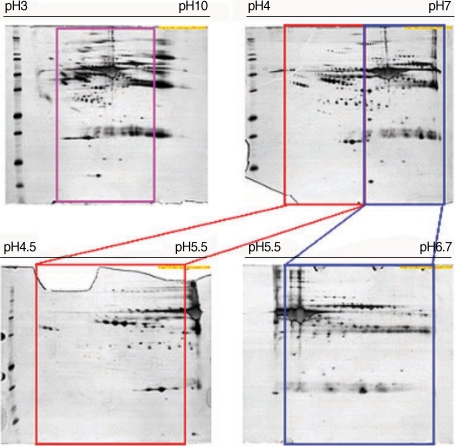
Fig. 2Comparison of 2-DE maps of plasma proteins from a normal healthy donor after depleting abundant plasma proteins. For these maps, 100 µg (pH range 3-10), 150 µg (4-7), 200 µg (4.5-5.5) and 200 µg (5.5-6.7) of plasma proteins were applied by in-gel rehydration to IPG strips after depleting 2, 6, or 20 abundant proteins in plasma using the ProteoExtract™ Albumin/IgG Removal kit, the multiple affinity removal column system (MARS®), or the ProteoPrep™ 20 column, respectively. For separation in the second dimension, 9-12% gradient SDS-PAGE was used. A, crude plasma sample; B, plasma sample after depleting 2 abundant proteins; C, plasma sample after depleting 6 abundant proteins; D, plasma sample after depleting 20 abundant proteins. The gels were visualized by silver staining and analyzed by the Melanie II program.
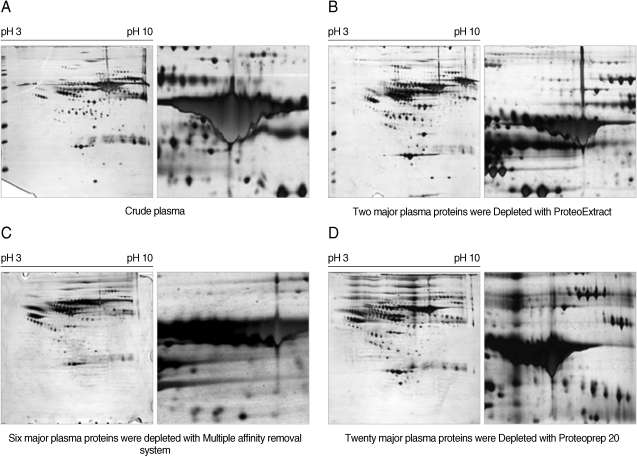
Fig. 3Comparison of the 2-DE maps of the plasma proteins from a healthy donor and a patient with vivax malaria. One hundred micrograms of whole plasma from a normal healthy individual (A) or a patient with vivax malaria (B) were separated on 2-DE gel electrophoresis with IPG strip on the pH range of 3-10 (nonlinear) and by 9-12% SDS-PAGE in the second dimension, respectively. The gels were stained by silver nitrate. Regions 1, 2, and 3 indicate β-chain, α-chain, and truncated α-chain proteins of haptoglobin, respectively, which were found to be significantly and consistently different between the plasma from patients with vivax malaria and normal healthy individuals.
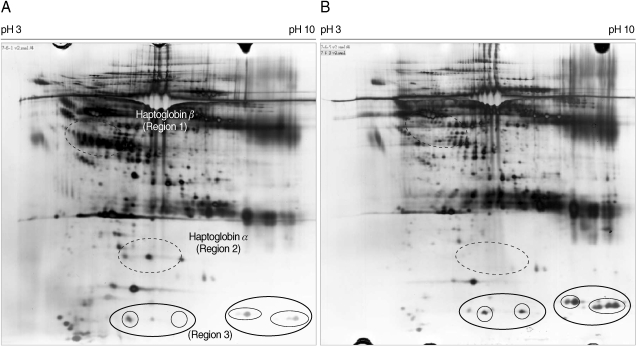
Fig. 4Magnified regions of the differentially-expressed haptoglobin in plasma from the normal healthy individuals and patients with vivax malaria. Each set shows gel regions 2 and 1 representing the α-chain and β-chain of haptoglobin, respectively. Normal regions 1 to 4 are show the magnified regions on each 2-DE gel with the plasmas from 4 healthy donors and 4 patients with vivax malaria. Arrows indicate the differentially expressed haptoglobin spots (A, α-chain of haptoglobin; B, β-chain of haptoglobin).
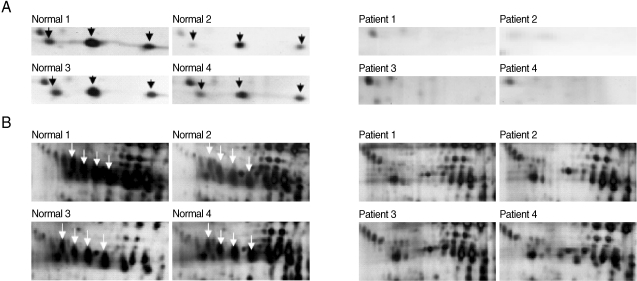
Fig. 5Magnified regions of the differentially expressed SAA protein spots on each 2-DE gel. Circled spots were identified as SAA proteins analyzed with 2-DE, SDS-PAGE and MALDI-TOF MS followed by identification with the search engine programs ProFound and Mascot. Normal denotes the plasma from a healthy donor and patients 1-5 denotes the plasma from 5 patients with vivax malaria.
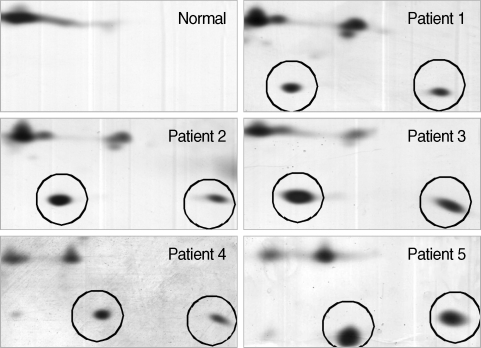
Fig. 6SAA protein in the plasma of vivax malaria patients detected by chemiluminescence. Lane 1-44, vivax patients; N1-4, normal healthy group. For immunoblot analysis, equal amounts (30 µg) of whole plasmas from individuals were analyzed with a commercially available anti-SAA polyclonal antibody followed by a goat antirabbit secondary antibody coupled to horseradish peroxidase. The immunoreactive proteins on the membrane were detected by chemilluminescence using the West-Save™ substrate onto X-ray film.