Abstract
We studied on the proteomic characteristics of Toxoplasma gondii KI-1 tachyzoites which were originally isolated from a Korean patient, and compared with those of the well-known virulent RH strain using 2-dimensional electrophoresis (2-DE), mass spectrometry, and quantitative real-time PCR. Two-dimensional separation of the total proteins isolated from KI-1 tachyzoites revealed up to 150 spots, of which 121 were consistent with those of RH tachyzoites. Of the remaining 29 spots, 14 showed greater than 5-fold difference in density between the KI-1 and RH tachyzoites at a pH of 5.0-8.0. Among the 14 spots, 5 from the KI-1 isolate and 7 from the RH strain were identified using MALDI-TOF mass spectrometry and database searches. The spots from the KI-1 tachyzoites were dense granule proteins (GRA 2, 3, 6, and 7), hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGRPTase), and uracil phosphoribosyltransferase (UPRTase). The spots from the RH strain were surface antigen 1 (SAG 1), L-lactate dehydrogenase (LDH), actin, chorismate synthase, peroximal catalase, hexokinase, bifunctional dihydrofolate reductase-thymidylate synthase (DHTR-TS), and nucleoside-triphosphatases (NTPases). Quantitative real-time PCR supported our mass spectrometric results by showing the elevated expression of the genes encoding GRA 2, 3, and 6 and UPRTase in the KI-1 tachyzoites and those encoding GRA 7, SAG 1, NTPase, and chorismate synthase in the RH tachyzoites. These observations demonstrate that the protein compositions of KI-1 and RH tachyzoites are similar but differential protein expression is involved in virulence.
-
Key words: Toxoplasma gondii, Korean Isolate-1 (KI-1), proteomics, quantitative real time PCR
INTRODUCTION
The obligate intracellular protozoan parasite
Toxoplasma gondii infects all warm-blooded vertebrates. Human toxoplasmosis is caused by the consumption of undercooked meat containing viable
T. gondii or the ingestion of oocysts shed from the feces of infected cats [
1]. Human toxoplasmosis is usually asymptomatic; however, cervical or occipital lymphadenophathy and ocular toxoplasmosis can occur in some patients. Congenital infection or reactivation in immunocompromised cases may lead to life-threatening encephalitis [
1].
Several strains of
T. gondii have been identified [
2]. In Korea,
T. gondii was isolated from 2 chorioretinitis patients [
3], 2 congenital patients [
4], and from the diaphragm of pigs [
5]; however, these isolates were neither maintained nor characterized. Recently,
T. gondii tachyzoites, designated as Korean Isolate-1 (KI-1), were successfully maintained in the laboratory [
6]. They proved to be highly virulent when inoculated into BALB/c mice and cultured in sarcoma 180 cells [
6], and were assigned to the genotype I clonal lineage using PCR-based restriction fragment length polymorphism (PCR-RFLP) analysis [
7].
Proteomic approaches for studying pathogenic parasites, including
T. gondii, are useful for understanding developmental stage-specific protein expression. Such analyses can provide information about specificity and virulence, and contribute to the development of therapeutic agents and an effective vaccine. A number of studies based on 2-dimensional electrophoresis (2-DE) and mass spectrometry have been performed on
T. gondii [
8-
12]. For example, Cohen et al. [
8] characterized global protein expression of the
T. gondii RH strain by 2-DE and matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, while Xia et al. [
11] analyzed the proteome of 3 strains of
Toxoplasma (ME49, GT1, and VEG) using 2-DE, gel liquid chromatography-linked tandem mass spectrometry, and multidimensional protein identification technology (MudPIT). However, proteomic information about geographic isolates of
T. gondii, including KI-1, is limited.
In Korea, the positive rate for anti-
Toxoplasma antibodies is approximately 7% [
13,
14] and several clinical cases have been reported [
15], but KI-1 is the only strain that has been maintained [
6]. The virulence and tissue culture characteristics of this unique isolate resemble those of type I strains [
7], but there is no information about its proteomic characteristics. Therefore, we compared the protein composition of tachyzoites of
T. gondii KI-1 with those of the RH strain using 2-DE and MALDI-TOF mass spectrometry.
MATERIALS AND METHODS
Total protein preparation from the parasites
T. gondii RH and KI-1 (provided by Dr. JY Chai, Seoul National University, Seoul, Korea) tachyzoites were proliferated in rat fibroblasts and collected by gradient centrifugation by Percoll, and then passed the 3 µm polycarbonate membranes (Whatman International Ltd., Kent, UK) to remove the host cells. Filtered tachyzoites were washed twice in PBS (pH 7.4) by centrifugation at 1,500 g for 20 min at 4℃, given a final wash in PBS, pelleted by centrifugation at 16,000 g for 10 min at 4℃, and stored in batches of 1 × 107 tachyzoites at -80℃. For total protein isolation, 1 × 107 tachyzoite pellets were disrupted in 100 µl of lysis buffer containing 8M urea, 4% CHAPS, 40 mM Tris and protease inhibitor cocktail (Roche Diagnostics GmBH, Mannheim, Germany) by rapid freezing and thawing using liquid nitrogen. The tachyzoites were further disrupted by sonication for 5 min at 4℃. Protein concentrations of the resulting supernatants were determined by a BCA Protein Assay kit (Pierce Biotechnology, Rockford, Illinois, USA).
2-DE
T. gondii proteins from 5 × 107 tachyzoites (approximately 100 µg) were mixed with rehydration buffer containing 9.5 M urea, 4% CHAPS, 50 mM DTT, 0.2% biolytes max, 40 mM Tris and bromophenol blue, to a final sample volume of 500 µl. Insoluble material was removed by centrifugation at 16,000 × g for 10 min at 4℃. For isoelectric focusing at first, the samples were separated using ReadyStrip IPG pH 3-10 non-linear strips (Bio-Rad Lab, Hercules, California, USA) or pH 5-8 linear strips according to the following conditions: 50 V for 12 hr, 250 V for 30 min, 4,000 V for 2 hr and 20,000 V for 5 hr at 20℃. After isoelectric focusing, the protein samples were redissolved in equilibration buffer (6 M urea, 2% SDS, 20% glycerol, 2% DTT, 2.5% iodoacetamide, and 0.375 M Tris pH 8.8) for 20 min. Discontinuous SDS-PAGE was performed next using 10% agarose gels, and the proteins were stained with silver nitrate. The gels were calibrated using weight and isoelectric point (pI) markers (Bio-Rad). The stained gels were scanned with Fluor-S™ Muti-Image (Bio-Rad). The molecular mass, pH, and density of the protein spots were analyzed using PDQuest™ 2-D Analysis Software (Bio-Rad).
MADLI-TOF mass spectrometry
Protein spots showing differential expression based on our 2-DE analysis were cut from the gels. The slices were homogenized, washed sequentially with 100 mM ammonium bicarbonate, 100 mM ammonium bicarbonate/50% acetonitrile, and 100% acetonitrile, and then dried. The isolated proteins were then trypsinized, mixed with 250 mM ammonium bicarbonate, concentrated, desalted using C18 Zip Tips (Millipore, Billerica, Massachusetts, USA), and analyzed by MALDI-TOF mass spectrometry using a Voyager™, DE-STR (PerSeptive Biosystems, Framingham, Massachusetts, USA) equipped with a nitrogen laser (337 nm, 3 ns pulse width, 3 Hz repetition rate). The peptide mass fingerprint data were used to search the following genomic and protein databases: Genpept translated nucleotide database (
http://www.ncbi.nlm.nih.gov/Entrez), Swissprot protein database, and pdbEST expressed sequence tag database (
http://www.ncbi.nlm.nih.gov/dbEST/index.html).
Total RNA was extracted from
T. gondii RH and KI-1 tachyzoites using Trizol (Invitrogen, Carlsbad, California, USA) reagent according to the manufacturer's instructions. cDNA was then synthesized using an RNA LA PCR Kit (Takara, Shiga, Japan). Real-time PCR was performed using iQ SyBr Green Supermix (Bio-Rad) polymerase and an iCycler (Bio-Rad) thermal cycler. The primer sets used for real-time PCR are described in
Table 1. The reaction conditions were 45 cycles of 95℃ for 30 sec, 60℃ for 30 sec, and 72℃ for 1 min. Gene expression was calculated by dividing the cycle threshold (C
T) value for each gene by that of the control,
GAPDH.
The data were analyzed using Student's t-test (2-tailed) or χ2 analysis. All experiments were carried out in duplicate or triplicate.
RESULTS
Proteins present differentially between the KI-1 and RH strains by 2-DE
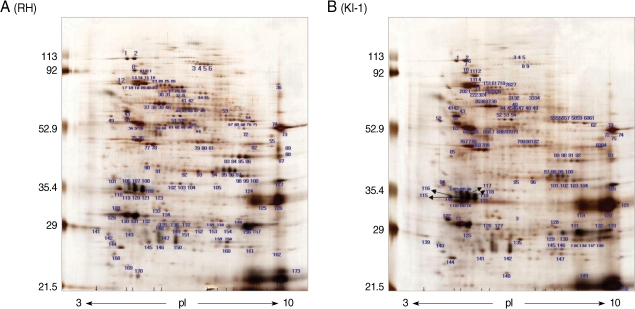
Two-dimensional gel electrophoresis of the total proteins from
T. gondii RH tachyzoites identified more than 163 protein spots within a pH range of 3-10 (
Fig. 1A). The spots were distributed mainly in the 82-85, 65-67, 53-54, 35-36, and 28-31 kDa ranges. Similar results were obtained in the 5-8 pH range (data not shown). In comparison, 2-DE of total proteins from
T. gondii KI-1 tachyzoites identified more than 150 protein spots (
Fig. 1B) of which 121 were identical in molecular weight and pH range to those of the RH strain. Fourteen spots (spot No. 34, 35, 47, 48, 79, 80, 81, 104, and 105 for the RH tachyzoites; spot No. 134, 138, 140, 144, and 148 for the KI-1 tachyzoites) within the 5-8 pH range showed a significant pH shift, difference in molecular weight, or density (
Table 2). Spots showing a 5-fold difference in density (mean density difference, 5.6 ± 0.7) between the RH and KI-1 strains were selected for further analysis by MALDI-TOF mass spectrometry.
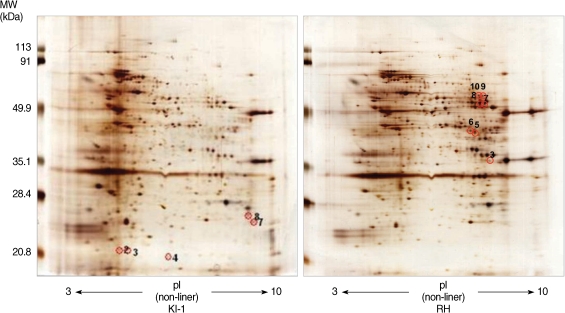
Among 14 spots showing a 5-fold difference in density between the RH and KI-1 strains, 5 protein spots from the KI-1 isolate and 7 from the RH strain were analyzed by MALDI-TOF mass spectrometry (
Fig. 2). The proteins expressed at a significantly higher level in the KI-1 tachyzoites were identified as dense granule proteins (GRA 2, 3, 6, and 7), hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGRPTase), and uracil phosphoribosyltransferase (UPRTase) (
Table 3). Those proteins expressed at a significantly higher level in the RH tachyzoites were identified as L-lactate dehydrogenase (LDH), actin, chorismate synthase, peroximal catalase, hexokinase, bifunctional dihydrofolate reductase-thymidylate synthase (DHTR-TS), nucleoside-triphosphatases (NTPases), and the major surface antigen of
T. gondii, surface antigen 1 (SAG 1) (
Table 4).
Differential expression of the proteins identified by 2-DE and MALDI-TOF mass spectrometry in the RH and KI-1 tachyzoites was confirmed using real-time PCR. Our results indicate that the expression levels of dense granule proteins (GRA 2, 3, and 6) and UPRTase were significantly higher in the KI-1 tachyzoites than in those of the RH strain. The expression levels of GRA 7, SAG 1, NTPases, and chorismate synthase were significantly higher in the RH tachyzoites than in the KI-1 strain (
Table 5). These data correspond to those from our MALDI-TOF analysis except for our results concerning the expression of GRA 7 and chorismate synthase. Real-time PCR analysis did not indicate a significant difference in the expression levels of HGRPTase, LDH, actin, peroximal catalase, hexokinase, and DHTR-TS between the RH and KI-1 tachyzoites (data not shown).
DISCUSSION
Most pathological changes due to T. gondii are caused by tachyzoites; thus, the identification and functional characterization of proteins expressed at this stage is important. T. gondii KI-1 tachyzoites isolated previously from a Korean patient were used in this study to investigate the differences in protein compositions between T. gondii RH and KI-1 tachyzoites. Silver staining following 2-DE revealed more than 150 and 167 protein spots from KI-1 and RH tachyzoites, respectively. Mass spectrometry and quantitative real-time PCR revealed that the expression levels of dense granule proteins were higher in the KI-1 tachyzoites, whereas SAG 1, NTPases, and chorismate synthase expression was higher in the RH strain. These observations demonstrate that the proteomic characteristics of KI-1 tachyzoites of T. gondii are different from those of the RH strain.
Toxoplasma isolates from humans and livestock are grouped into 3 clonal lineages (types I, II, and III), which can be discriminated by their virulence in mice [
2]: strains classified as type I are highly virulent, while types II and III are less virulent.
T. gondii strain KI-1 was classified as a type I regional variant [
7], but until now there have been no reports describing the antigenic characteristics of its tachyzoites. Proteomics has become a key tool in the investigation of proteins by 2-DE and mass spectrometry [
16]. In this study, 2-DE was used to compare the protein composition of KI-1 and RH tachyzoites. To minimize host cell contamination, we used filtration through 3-µm polycarbonate membranes, differential centrifugation, and microscopic confirmation before protein solubilization. Two-dimensional electrophoresis of total proteins extracted from
T. gondii KI-1 tachyzoites identified more than 150 protein spots in the 3-10 pH range, which is similar to the number of spots detected for the RH strain. Nischik et al. [
12] compared the proteomes of the virulent and attenuated
T. gondii BK strain and found approximately 200 matching proteins. Cohen et al. [
8] analyzed a map of
T. gondii tachyzoites produced by 2-DE and found more than 1,000 polypeptides that were reproducibly separated at a high resolution by 2-DE in the pH ranges of 4-7 and 6-11. Dlugonska et al. [
9] resolved more than 200 spots of
T. gondii tachyzoite lysates, and identified eleven excretory-secretory dense granule proteins that contained B cell epitopes, as well as 2 containing T cell epitopes. Ma et al. [
10] reported that 1,227 protein spots of
T. gondii soluble tachyzoite antigen were fractionated by 2-DE at a pH range of 3-10, and they identified 426 by mass spectrometric analysis and database searches. Xia et al. [
11] used 3 complementary approaches (2-DE, gel-liquid chromatography linked tandem mass spectrometry, and MudPIT), and identified nearly one-third (2,252) of all the predicted proteins of
T. gondii, with 2,477 intron-spanning peptides providing evidence for correct splice site annotation. Proteomics study is a powerful tool to identify the protein composition; however, careful interpretation of the data is required. Consideration should be made with regard to standardizing sample conditions, sample preparations, experimental conditions and experimental procedures.
Protein spots showing a greater than 5-fold difference in density between
T. gondii KI-1 and RH tachyzoites were excised from the gels and further characterized by MALDI-TOF mass spectrometry and quantitative real-time PCR. The greater expression of GRA 2, 3, and 6 and UPRTase found in the KI-1 tachyzoites confirm previous reports showing that the KI-1 tachyzoite is a virulent local isolate [
7]. Dense granule protein expression was higher in the KI-1 tachyzoites than in the RH tachyzoites;
T. gondii GRAs may participate in the modification of parasitophorous vacuoles (PVs) and the PV membrane, which are required for the maintenance of intracellular parasitism in almost all nucleated host cells [
17]. GRA 1-5, 7, and 8; microneme protein 5; NTPase-I; catalase; and actin are known to play a role in the virulence of
T. gondii tachyzoites [
12]. GRA 7, SAG 1, NTPases and chorismate synthase expression was elevated in the RH strain compared to the KI-1 isolates.
T. gondii SAG 1 may play a crucial role in immune modulation or virulence attenuation. NTPase is abundantly released from dense granules of
T. gondii; 2 known isoforms, NTPase-I and NTPase-II, have been identified in tachyzoites. Interestingly, the gene encoding NTPase-II has been found in all strains of
T. gondii, whereas the gene encoding NTPase-I is restricted to virulent strains. These reports suggest that the virulent characteristics of KI-1 tachyzoites are different from those of the RH strain, although they are both type I (virulent) strains [
7].
T. gondii has various strains and different developmental stages, each of which contains unique antigens. In this study, we compared the proteomic differences between T. gondii tachyzoites of the RH strain and the Korean KI-1 isolate using 2-DE, mass spectrometry, and quantitative real-time PCR. Our results indicate that the protein composition of KI-1 tachyzoites is similar to that of the RH strain, but that differential protein expression is involved in virulence. In this study, the identification of low-density proteins was limited; future analyses of the T. gondii proteome by more advanced technical applications will likely reveal other characteristic antigens.
ACKNOWLEDGEMENTS
This work was supported by a National Research Foundation of Korea Grant funded by the Korean Government (2009-00-68706), the Korea Science & Engineering Foundation through the Infection Signaling Network Research Center (R13-2007-020-01000-0) and the 2007 Chungnam National University Hospital Research Fund.
References
- 1. Dodds EM. Toxoplasmosis. Curr Opin Ophthalmol 2006;17:557-561.
- 2. Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: Correlation of parasite genotype with human diseases. J Infect Dis 1995;172:1561-1566.
- 3. Choi JS, Choi CS, Soh CT. Isolation of Toxoplasma gondii from congenital and acquired chorioretinitis cases. Yonsei Rep Trop Med 1980;11:39-42.
- 4. Chung KS, Kung RN, Chung KS, Kim PK, Yun DJ, Soh CT. Congenital toxoplasmosis. Yonsei Med J 1980;21:62-74.
- 5. Choi WY. The isolation of Toxoplasma gondii from pork and the dye test fo swine sera. J Catholic Med College 1969;16:229-235.
- 6. Chai JY, Lin A, Shin EH, Oh MD, Han ET, Nan HW, Lee SH. Laboratory passage and characterization of an isolate of Toxoplasma gondii from an ocular patient in Korea. Korean J Parasitol 2003;41:147-154.
- 7. Lin A, Shin EH, Kim TY, Park JH, Guk SM, Chai JY. Genetic characteristics of the Korean isolate KI-1 of Toxoplasma gondii. Korean J Parasitol 2005;43:27-32.
- 8. Cohen AM, Rumpel K, Coombs GH, Wastling JM. Characterization of global protein expression by two-dimensional electrophoresis and mass spectrometry: proteomics of Toxoplasma gondii. Int J Parasitol 2002;32:39-51.
- 9. Dlugonska H, Dytnerska K, Reichmann G, Stachelhaus S, Fischer HG. Towards the Toxoplasma gondii proteome: position of 13 parasite excretory antigens on a standardized map of two-dimensionally separated tachyzoite proteins. Parasitol Res 2001;87:634-637.
- 10. Ma GY, Zhang JZ, Yin GR, Zhang JH, Meng XL, Zhao F. Toxoplasma gondii: proteomic analysis of antigenicity of soluble tachyzoite antigen. Exp Parasitol 2009;122:41-46.
- 11. Xia D, Sanderson SJ, Jones AR, Prieto JH, Yates JR, Bromley E, Tomley FM, Lal K, Sinden RE, Brunk BP, Roos DS, Wastling JM. The proteome of Toxoplasma gondii: integration with the genome provides novel insights into gene expression and annotation. Genome Biol 2008;9:R116.
- 12. Nischik N, Schade B, Dytnerska K, Długońska H, Reichmann G, Fischer HG. Attenuation of mouse-virulent Toxoplasma gondii parasites is associated with a decrease in interleukin-12-inducing tachyzoite activity and reduced expression of actin, catalase and excretory proteins. Microbes Infect 2001;3:689-699.
- 13. Lee YH, Noh HJ, Hwang OS, Lee SK, Shin DW. Seroepidemiological study of Toxoplasma gondii infection in the rural area Okcheon-gun, Korea. Korean J Parasitol 2000;38:251-256.
- 14. Yang HJ, Jin KN, Park YK, Hong SC, Bae JM, Lee SH, Choi HS, Hwang HS, Chung YB, Lee NS, Nam HW. Seroprevalence of toxoplasmosis in the residents of Cheju island, Korea. Korean J Parasitol 2000;38:91-93.
- 15. Lee JA, Kim DH, Kim YK, Chung EH, Choi JH, Lee HJ, Chi JG, Chai JY, Lee YH. Two cases of congenital toxoplasmosis diagnosed by polymerase chain reaction. Infect Chemother 2003;35:45-52.
- 16. Weiss LM, Fiser A, Angeletti RH, Kim K. Toxoplasma gondii proteomics. Expert Rev Proteomics 2009;6:303-313.
- 17. Nam HW. GRA proteins of Toxoplasma gondii: maintenance of host-parasite interactions across the parasitophorous vacuolar membrane. Korean J Parasitol 2009;47(suppl):S29-S37.
Fig. 1Separation of total proteins extracted from T. gondii RH (A) and KI-1 (B) tachyzoites by 2-DE. KI-1, isolated from the blood of an ocular patient in Korea; RH, virulent strain of T. gondii. The proteins were focused to their isoelectric points using 17-cm ReadyStrip™ IPG pH 3-10 non-linear gradient strips. Each panel is representative of 3 independent experiments.

Fig. 2Protein spots identified by 2-DE showing a higher than 5-fold difference in density between KI-1 and RH gels were analyzed by MALDI-TOF mass spectrometry. In the KI-1 gel, identification numbers (ID#) 2, 3, 4, 7, and 8 were analyzed, while in the RH gel, ID# 3, 5, 6, 7, 8, 9, and 10 were analyzed. Each panel is representative of 3 independent experiments.

Table 1.Primer sequences for quantitative real time PCR
Table 1.
|
Protein |
Primer sequence |
Product size (bp) |
|
GRA2 |
Forward |
AGAGGCAACAAGAGCCAGAA |
200 |
|
Reverse |
TTCTTTGGCCACCTTGAAAC |
|
|
GRA3 |
Forward |
GGAAGATGATCAGGCTCTCG |
197 |
|
Reverse |
GATGGTCGCTCTCTCCAGTC |
|
|
GRA6 |
Forward |
GGGCACAAGACGACGTTATT |
203 |
|
Reverse |
TATTGCCTGCATCATTTCCA |
|
|
GRA7 |
Forward |
GTGCCGGAACTAACAGAGGA |
200 |
|
Reverse |
GATTCAGGCACCTCTTGCTC |
|
|
HGXPRTasea
|
Forward |
GGTAAATGTCCCGTCGAGAA |
197 |
|
Reverse |
GGAGTTGGGTCGAGAGAACA |
|
|
UPRTaseb
|
Forward |
CAGGACAGGCACACTCGTTA |
203 |
|
Reverse |
TGCTTCCTTCTCGTTCATCA |
|
|
LDHc
|
Forward |
ATTGCTGACACGAACGTGAG |
199 |
|
Reverse |
CAAGCGGGCAGTACTTCTTC |
|
|
Chorismate synthase |
Forward |
GGCTTGGAACAAGAAACTGC |
201 |
|
Reverse |
GCAACCACTGTGTTGAGGAA |
|
|
DHFR-TSd
|
Forward |
GCACACGTCTGCAACCTAAA |
197 |
|
Reverse |
TAGCCCACGACCTCAAAATC |
|
|
SAG1 |
Forward |
TGTCAAGTTGTCTGCGGAAG |
197 |
|
Reverse |
TGGCACCATTATCACTCGAA |
|
|
Actin |
Forward |
AGGATCTGTACGGCAACGTC |
201 |
|
Reverse |
TGATCCACATCTGCTGGAAG |
|
|
NTPasee-I |
Forward |
GAGTGGACGCTGACACTGAA |
192 |
|
Reverse |
GGTCAATGCTACGGCCTTTA |
|
|
NTPase-II |
Forward |
TTGCCATTTTGGACTCACAG |
205 |
|
Reverse |
CGTGTCCAGAAAGGATTGGT |
|
|
Hexokinase |
Forward |
CTGATGGATAGGTCCGGAGA |
193 |
|
Reverse |
CAAGCAACACTGCCACATCT |
|
|
Peroximal catalase |
Forward |
CTCATATGGTGCCAGGGATT |
200 |
|
Reverse |
CAGCTGAGACCCTTTGTTCC |
|
|
GAPDH |
Forward |
CCAAGCACGCCATTATTTCT |
207 |
|
Reverse |
TCGCTGCATGTACGGTAGTC |
|
Table 2.Protein spots identified by 2-DE showing a higher than 5-fold difference in density between KI-1 and RH gels
Table 2.
|
Spot No. |
pH |
Molecular weight (kDa) |
Type of tachyzoite |
|
34 |
5.8 |
74 |
RH |
|
35 |
6.0 |
74 |
RH |
|
47 |
5.7 |
65 |
RH |
|
48 |
5.9 |
65 |
RH |
|
79 |
5.8 |
48 |
RH |
|
80 |
5.8 |
48 |
RH |
|
81 |
5.9 |
48 |
RH |
|
104 |
5.8 |
36 |
RH |
|
105 |
6.9 |
36 |
RH |
|
134 |
8.7 |
26 |
KI-1 |
|
138 |
8.7 |
26 |
KI-1 |
|
140 |
4.2 |
26 |
KI-1 |
|
144 |
4.7 |
24 |
KI-1 |
|
148 |
6.0 |
23 |
KI-1 |
Table 3.Identification of KI-1 tachyzoites protein spots showing more than 5 times density difference in 2-DE analysis between KI-1 and RH gels
Table 3.
|
ID |
Protein MW Da/pI |
Masses matched (Sequence covered; %) |
Protein name |
|
KI-1 #2 |
25858/5.1 |
5 (30) |
GRA7 precursor |
|
24030/5.5 |
3 (23) |
GRA6 |
|
26386/5.9 |
1 (5) |
HGXPRTase |
|
KI-1 #3 |
25858/5.1 |
16 (4.2) |
GRA7 precursor |
|
KI-1 #4 |
25858/5.1 |
3 (15) |
GRA7 precursor |
|
27647/6.6 |
2 (9) |
UPRTase |
|
24030/5.5 |
3 (13) |
GRA6 |
|
26386/5.9 |
2 (8) |
HGXPRTase |
|
23644/4.5 |
1 (4) |
GRA3 precursor |
|
KI-1 #7 |
19843/9.0 |
2 (15) |
GRA2 precursor |
|
KI-1 #8 |
19843/9.0 |
1 (4) |
GRA2 precursor |
Table 4.Identification of RH strain protein spots showing more than 5 times density difference in 2-DE analysis between KI-1 and RH gels
Table 4.
|
ID |
Protein MW Da/pI |
Masses matched (Sequence covered; %) |
Protein name |
|
RH #3 |
34829/8.3 |
1 (7) |
SAG1 precursor |
|
35307/6.5 |
1 (7) |
LDH |
|
RH #5 |
41908/5.0 |
1 (3) |
Actin |
|
RH #6 |
41908/5.0 |
2 (5) |
Actin |
|
35307/6.5 |
1 (5) |
LDH |
|
RH #7 |
58145/7.9 |
2 (4) |
Chorismate synthase |
|
57271/6.7 |
1 (2) |
Peroxisomal catalase |
|
RH #8 |
51468/5.9 |
1 (1) |
Hexokinase |
|
57271/6.7 |
2 (4) |
Peroxisomal catalase |
|
58145/7.9 |
2 (3) |
Chorismate synthase |
|
RH #9 |
68752/6.8 |
2 (3) |
DHFR-TS |
|
58145/7.9 |
2 (5) |
Chorismate synthase |
|
51468/5.9 |
1 (2) |
Hexokinase |
|
69586/5.7 |
2 (5) |
NTPase-II precursor |
|
65763/6.5 |
3 (7) |
Putative NTPase |
|
69160/6.0 |
1 (3) |
NTPase-I precursor |
|
RH #10 |
58145/7.9 |
4 (5) |
Chorismate synthase |
|
51468/5.9 |
2 (4) |
Hexokinase |
|
65763/6.5 |
2 (2) |
Putative NTPase |
|
68752/6.8 |
1 (1) |
DHFR-TS |
|
69160/6.0 |
1 (1) |
NTPase-I precursor |
|
69586/5.7 |
1 (1) |
NTPase-II precursor |
Table 5.Real time PCR of proteins showed different expression in MALDI-TOF analysis
Table 5.
|
Protein |
CT valuea
|
P-value |
|
KI-1 |
RH |
Fold-difference (KI-1/RH) |
|
GRA2 |
0.741 ± 0.028 |
0.683 ± 0.003 |
1.085 |
0.024448 |
|
GRA3 |
1.123 ± 0.019 |
0.941 ± 0.015 |
1.193 |
0.000209 |
|
GRA6 |
0.954 ± 0.009 |
0.823 ± 0.023 |
1.159 |
0.000758 |
|
GRA7 |
0.815 ± 0.023 |
0.889 ± 0.039 |
0.917 |
0.04912 |
|
SAG1 |
0.721 ± 0.015 |
0.781 ± 0.017 |
0.923 |
0.010799 |
|
NTPaseI |
0.826 ± 0.005 |
0.898 ± 0.044 |
0.920 |
0.047986 |
|
NTPaseII |
0.769 ± 0.016 |
0.844 ± 0.034 |
0.911 |
0.026487 |
|
UPRTase |
1.697 ± 0.0141 |
1.499 ± 0.030 |
1.132 |
0.000489 |
|
Chorismate synthase |
1.143 ± 0.056 |
1.134 ± 0.0441 |
1.008 |
0.009063 |