Abstract
The ascarids, Toxocara canis and Toxascaris leonina, are probably the most common gastrointestinal helminths encountered in dogs. In order to understand biological differences of 2 ascarids, we analyzed gene expression profiles of female adults of T. canis and T. leonina using CLC Genomics Workbench, and the results were compared with those of free-living nematode Caenorhabditis elegans. A total of 2,880 and 7,949 ESTs were collected from T. leonina and T. canis, respectively. The length of ESTs ranged from 106 to 4,637 bp with an average insert size of 820 bp. Overall, our results showed that most functional gene annotations of 2 ascarids were quite similar to each other in 3 major categories, i.e., cellular component, biological process, and molecular function. Although some different transcript expression categories were found, the distance was short and it was not enough to explain their different lifestyles. However, we found distinguished transcript differences between ascarid parasites and free-living nematodes. Understanding evolutionary genetic changes might be helpful for studies of the lifestyle and evolution of parasites.
-
Key words: Toxascaris leonina, Toxocara canis, Caenorhabditis elegans, Gene ontology, CLC genomics workbench
INTRODUCTION
The ascarids,
Toxocara canis and
Toxascaris leonina, are probably the most common gastrointestinal helminths encountered in dogs. These ascarids have similar aspects; their genera are included in the Family Ascarididae and their morphological features are difficult to distinguish from one another by naked eyes of a non-professional person [
1]. However, they also exhibit some different biological features. Prenatal transmission assures that virtually all puppies are born infected with
T. canis [
1,
2]. This transplacental transmission is observed in the life cycle of other
Toxocara sp.; it is probably the major route of infection for kittens with
T. cati.
T. canis can cause a serious disease in puppies; however,
T. leonina is generally less pathogenic because they have no prenatal transmission ability. In a young host,
T. canis can migrate to several internal organs (intestine-lung-trachea-intestine) during their juvenile stage for maturation into adult worms; however,
T. leonina larvae can mature to be adults in the intestinal cavity without tissue migration [
1]. Finally,
T. canis can grow into an adult only in canid hosts; however,
T. leonina can grow to be an adult in both canid and felid hosts [
1].
In order to characterize the biology, pathogenesis, and differential diagnosis of the 2 genera, many genetic variation studies have been reported [
3-
6]. Most studies dealt with genetic and serologic characteristics for the diagnosis of infection with 2 parasites. The parasites have similar antigenic characteristics, and
T. canis-infected host serum can be reacted with
T. leonina antigens [
3]. However, sequences of
cox2 in mitochondrial DNA and ITS-2 region of ribosomal DNA of
Toxocara and
Toxascaris differed significantly from one another [
4,
7]. However, no study comparing functional gene annotation profiles between
T. canis and
T. leonina has been reported.
Functional genomic strategies, such as the expressed sequence tag (EST), are a powerful tool for identification of large numbers of genes [
8]. EST analysis is not only one of the more effective methods for gene discovery, gene expression profiling, and functional genome studies, but also one of the more efficient methods for identification of differential genes [
8]. EST analyses of
T. canis and
T. leonina have been reported, respectively [
8-
10]. An efficient functional annotation of DNA or protein sequences is a major requirement for successful application of these approaches as functional information on gene products is often the key for interpretation of experimental results [
11]. Recently, a new functional annotation program, Blast2GO according to Gene Ontology (GO) criteria has been introduced; it is a powerful tool for comparison of functional gene expression profiles between 2 genetically similar organisms [
12-
14]. The GO project provides ontology of defined terms representing properties of gene products. The ontology covers 3 domains; cellular component (parts of a cell or its extracellular environment), molecular function (elemental activities of a gene product at the molecular level, such as binding or catalysis), and biological process (operations or sets of molecular events with a defined beginning and end, pertinent to functioning of integrated living units: cells, tissues, organs, and organisms) (
http://www.geneontology.org).
The aim of this study was to understand parasite-specific functional gene expression profiles of ascarids, and difference of gene expression profiles between the 2 ascarids. We analyzed gene expression profiles of female adults of T. canis and T. leonina using CLC Genomics Workbench and the results were compared with those of a free-living nematode species, Caenorhabditis elegans. Conducts of functional genomic studies give to us understanding of parasitism, development, metabolism, and reproduction of these parasites.
MATERIALS AND METHODS
Parasite
Adult
T. leonina and
T. canis female worms were collected from naturally infected dogs in the South Korea (=Korea). The worms were classified in accordance with the morphological features of the eggs and adults [
1].
For construction of cDNA libraries, the total RNAs of adult T. leonina and T. canis female worms were purified according to the protocol of QIAzol (Qiagen, Valencia, California, USA). Each of 5 µg of poly A+ mRNA was prepared from the total RNA using the absolute mRNA purification kit (Stratagene, La Jolla, California, USA). The cDNA libraries were constructed using the ZAP Express cDNA synthesis kit according to the manufacturer's instructions (Stratagene, San Diego, California, USA).
cDNA sequencing and analysis of ESTs
The sequence files obtained from cDNA of
T. leonina and
T. canis were initially submitted for pre-processing (Insilicogen, Suwon, Korea). This process was trimmed with sequences of quality scores 0.05 and ambiguous nucleotides. Each vector sequence was identified and trimmed off using the UniVec. EST sequences shorter than 100 bp were discarded. EST sequences were assembled and clustered using CLC Genomics Workbench. Finally, clusters and singletons were analyzed for annotation using a homology search engine, local BLAST [
15,
16] against the NCBI database (National Library of Medicine, Bethesda, Maryland, USA). Significant matches were determined when expectation value was less than 1
e-4 with previously reported genes in the NCBI database. The function of ESTs was predicted through Clusters of Orthologous Groups of proteins (COGs) analysis [
17]. Briefly, transcription profiles were created for the
T. canis and
T. leonina library using transcripts that matched sequences characterized in 2 other organisms;
C. elegans 23,906 protein sequence and
Ascaris lumbricoides 106,573 nucleotide. These were subjected to further BLAST analysis in order to obtain this information and were manually scrutinized for determination of the most meaningful annotation for each EST within the gene ontology (GO) scheme (
Supplementary Table 1). Results of GO analysis of 2 organisms were compared to each other, and to those of
C. elegans in AmiGO (
http://amigo.geneontology.org).
RESULTS
EST analysis
A summary of library data is shown in
Table 1. From the randomly selected plasmids containing DNA inserts, a total of 2,880 and 7,949 ESTs of
T. leonina and
T. canis adult female worm cDNA library were sequenced, respectively. The length of ESTs ranged from 106 to 4,637 bp with an average insert size of approximately 820 bp.
Using the CLC Genomics Workbench (CLCbio, Seoul, Korea), UniScripts were compared to the annotations in the Gene Ontology Consortium of C. elegans and common worm. GO terms were obtained for 1,178 of these UniScripts. Function annotation analysis was also used for determination of EC number and for identification of COGs and GO-annotated proteins. For comparison of biological and functional differences, we differentiated ESTs as cellular component, biological processing, and molecular function, and Level 2 GO terms for enriched UniScripts comprised of 5 or more ESTs. As shown in the results, types of proteins expressed in T. leonina and T. canis female adult worms did not differ significantly from each other based on GO categories.
Cellular component level 2 GO terms analyses of 3 organisms
We classified the transcripts according to 16 cellular component categories (cell, cell junction, cell part, extracellular matrix, extracellular matrix part, extracellular region, extracellular region part, macromolecular complex, membrane, membrane part, membrane-enclosed lumen, nucleoid, organelle, organelle part, synapse, and synapse part). In the cellular component category, we found most expressed genes localized in cells (20% and 21%, respectively), cell part (20% and 21%), organelle (17% and 16%), and organelle part (11% and 11%) of
T. leonina and
T. canis (
Fig. 1). Some difference was observed within the extracellular region subcategory, which comprised of 5.9% of predicted
T. canis ESTs compared with 3.7% of predicted
T. leonina ESTs. Extracellular regions of subcategories were consistent with extracellular space and extracellular matrix proteins, collagen, and collagen type V protein. However, in level 2 GO analyses of
C. elegans, the genes showing major expression were categorized as membrane and membrane part (meaning of 'part' is that the classified genes are related to the category, or their ancestor genes are related to the category). The proportion of cell, cell part, organelle, and organelle part categories of
C. elegans was lower than those of parasites.
The biological process-related transcripts were classified according to 23 categories (biological adhesion, biological regulation, cell killing, cellular component organization or biogenesis, cellular process, developmental process, establishment of localization, growth, immune system process, localization, locomotion, metabolic process, multi-organism process, multicellular organisms process, negative regulation of biological process, positive regulation of biological process, regulation of biological process, reproduction, reproductive process, response to stimulus, rhythmic process, signaling, and single organism process). Most biological process level 2 GO terms associated with
T. leonina and
T. canis of ESTs were also similar to each other. The most highly expressed biological process gene subcategories of 2 parasites were cellular process, developmental process, metabolic process, and multicellular organismal process (
Fig. 2). However, transcripts of free-living nematodes were the most classified as single organism process category, but those of parasites were not classified.
Biological process-related transcripts were classified according to 16 categories (antioxidant activity, binding, catalytic activity, cannel regulator activity, electron carrier activity, metallochaperone activity, molecular transducer activity, morphogen activity, nutrient reservoir activity, protein tag, receptor activity, receptor regulator, structure molecule activity, translation regulator activity, and transporter activity). The most highly expressed were binding (45% and 48%, respectively), catalytic activity proteins (28% and 26%), and structural molecule activity (13% and 11%) of
T. leonina and
T. canis (
Fig. 3). However, in
C. elegans, the most highly expressed was the electron carrier activity category (35%), on the other side, those of the parasites were consistent with a low proportion (2 and 3%). Morphogen activity, nutrient reservoir activity, and receptor activity of
C. elegans were higher than those of the 2 parasites.
DISCUSSION
Functional annotation of sequence data is a key requirement for successful generation of functional genomics in biological researches. In this study, we evaluated functional annotation of ESTs of T. leonina, T. canis, and C. elegans using the CLC Genomics Workbench program, in order to find some genetic difference for interpretation of biological differences between 2 ascarids and a free-living nematode.
Instead of our expectation, overall, our results showed that most functional gene annotations of 2 ascarids were quite similar to one another. We were not surprised by this result because they are included in the same Family Ascarididae. In particular, comparative analysis of biological process level 2 GO showed similar profiles between the 2 ascarids. Most ascarids, including
T. canis, require internal organ migration periods for growth to the adult. However,
T. leonina can grow to the adult without any migration period [
1].
T. leonina can grow to be adults without any larval migration; this fact suggests several hypotheses. One is that molting and development of
T. leonina can occur easily, compared with
T. canis; in other words,
T. leonina might have more abundant proteins related to molting and development. The other is that
T. leonina does not require certain molecules that may be released from host tissues (lung, airway tract, blood, etc.) and may be critical for molting and development of other ascarids. In the molecular function category, structural molecule activity proteins showed greater expression in
T. leonina than in
T. canis. Structural molecule activity category, including motor proteins (actin, myosins, and dyneins), are important to molting and development as well as to movement [
18]. Ubiquitin, heat-shock protein 70, α-actin, and β-actin are overexpressed in the abdominal muscle of marine shrimp
Litopenaeus vannamei during the molting cycle [
18]. This result suggested that perhaps
T. leonina have more moving activity and ability to molt easily compared with
T. canis. We cautiously suggested that these abilities articulated that molting and development of
T. leonina could occur without internal organ migration during their life cycle. However, our analyzed data were obtained from adult female worms, and the data were not sufficiently complete to explain the hypothesis.
However, we found some difference between free-living nematodes and parasites. Membrane and membrane part categories were composited with intrinsic to, extrinsic to, and integral to membrane proteins. These proteins were more enriched in free-living nematodes than in parasites (
Fig. 1). This result suggested that membrane complex might have been uncomplicated during evolutionary periods of parasites. In molecular function categories, we found a significantly different proportion of subcategories between parasites and free-living nematodes. In particular, the electron carrier activity category showed higher expression in
C. elegans than in the other 2 parasites. This category included hydrolase activity, transferase activity, and oxidoreductase activity related proteins, which were closely related to the metabolic system. These results suggested that free-living nematodes have a more complex metabolic system than parasitic nematodes. Most digestive, metabolite systems of parasites might have degenerated, compared with those of their ancestors during evolutionary periods [
1]. The most highly expressed transcripts of
C. elegans were classified as single-organism process category in Biological Process level 2 GO terms, but a few transcripts of parasites were classified in this category. However, some subcategories in the single organism process category were also classified in the cellular process category. In order to identify the transcripts in this category, we have to classify the transcripts according to the subcategory level in the single-organism process and cellular process category.
In this study, although no significant difference was observed between T. leonina and T. canis female adults to explain differences in their life cycle, we observed difference of transcript expression between parasites and free-living nematodes. Thus, it might be helpful to understand evolutionary genetic changes of parasites. Analysis of detailed functional individual gene study is needed in order to understand parasite-specific transcript expression patterns.
Pusan National University Hospital, Yangsan2011-04
Notes
-
The authors have no financial conflict of interest.
ACKNOWLEDGMENT
This study was supported by Medical Research Institute Grant (2011-04), Pusan National University Hospital, Yangsan, Korea.
Supplementary Material
References
- 1. Roberts LS, Schmidt GD, Janovy J. Foundations of Parasitology. Boston, USA. McGraw-Hill Higher Education; 2009.
- 2. Parsons JC. Ascarid infections of cats and dogs. Vet Clin North Am Small Anim Pract 1987;17:1307-1339.
- 3. Cuellar C, Fenoy S, Guillen JL. Cross-reactions of sera from Toxascaris leonina and Ascaris suum infected mice with Toxocara canis, Toxascaris leonina and Ascaris suum antigens. Int J Parasitol 1995;25:731-739.
- 4. Li MW, Lin RQ, Song HQ, Sani RA, Wu XY, Zhu XQ. Electrophoretic analysis of sequence variability in three mitochondrial DNA regions for ascaridoid parasites of human and animal health significance. Electrophoresis 2008;29:2912-2917.
- 5. Mikaeili F, Kia EB, Sharbatkhori M, Sharifdini M, Jalalizand N, Heidari Z, Zarei Z, Stensvold CR, Mirhendi H. Comparison of six simple methods for extracting ribosomal and mitochondrial DNA from Toxocara and Toxascaris nematodes. Exp Parasitol 2013;134:155-159.
- 6. Pawar RM, Lakshmikantan U, Hasan S, Poornachandar A, Shivaji S. Detection and molecular characterization of ascarid nematode infection (Toxascaris leonina and Toxocara cati) in captive Asiatic lions (Panthera leo persica). Acta Parasitol 2012;57:67-73.
- 7. Fogt-Wyrwas R, Mizgajska-Wiktor H, Pacon J, Jarosz W. Intraspecific variation between the ITS sequences of Toxocara canis, Toxocara cati and Toxascaris leonina from different host species in south-western Poland. J Helminthol 2012;87:432-442.
- 8. Zhou R, Xia Q, Huang H, Lai M, Wang Z. Construction of a cDNA library from female adult of Toxocara canis, and analysis of EST and immune-related gene expressions. Exp Parasitol 2011;129:120-126.
- 9. Cho MK, Lee KH, Lee SJ, Kang SW, Ock MS, Hong YC, Lee YS, Yu HS. Identification of host immune regulation candidate genes of Toxascaris leonina by expression sequenced tags (ESTs) analysis. Vet Parasitol 2009;164:242-247.
- 10. Maizels RM, Tetteh KK, Loukas A. Toxocara canis: genes expressed by the arrested infective larval stage of a parasitic nematode. Int J Parasitol 2000;30:495-508.
- 11. Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 2008;36:3420-3435.
- 12. Conesa A, Gotz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008;2008:619832.
- 13. Vieira PM, Coelho AS, Steindorff AS, de Siqueira SJ, Silva Rdo N, Ulhoa CJ. Identification of differentially expressed genes from Trichoderma harzianum during growth on cell wall of Fusarium solani as a tool for biotechnological application. BMC Genomics 2013;14:177.
- 14. Fernandez P, Soria M, Blesa D, DiRienzo J, Moschen S, Rivarola M, Clavijo BJ, Gonzalez S, Peluffo L, Principi D, Dosio G, Aguirrezabal L, Garcia-Garcia F, Conesa A, Hopp E, Dopazo J, Heinz RA, Paniego N. Development, characterization and experimental validation of a cultivated sunflower (Helianthus annuus L.) gene expression oligonucleotide microarray. PLoS One 2012;7:e45899.
- 15. Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A 1990;87:5509-5513.
- 16. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403-410.
- 17. Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 2000;28:33-36.
- 18. Renfranz PJ, Blankman E, Beckerle MC. The cytoskeletal regulator zyxin is required for viability in Drosophila melanogaster. Anat Rec (Hoboken) 2010;293:1455-1469.
Fig. 1Comparative analysis of Cellular Component Level 2 GO terms associated with C. elegans, T. canis, and T. leonina enriched UniScripts. Cellular Component Level 2 GO terms were derived for stage-enriched UniScripts using CLC Genomics Workbench. Marked differences were observed in the types of proteins expressed in C. elegans, T. canis, and T. leonina, most clearly in the cell and organelle subcategories.

Fig. 2Comparative analysis of Biological Process level 2 GO terms associated with C. elegans, T. canis, and T. leonina enriched UniScripts. Biological Process Level 2 GO terms were derived for stage-enriched UniScripts using CLC Genomics Workbench. Differences in the types of proteins expressed in C. elegans, T. canis, and T. leonina were apparent in several subcategories, including cellular, metabolic, and developmental processes.
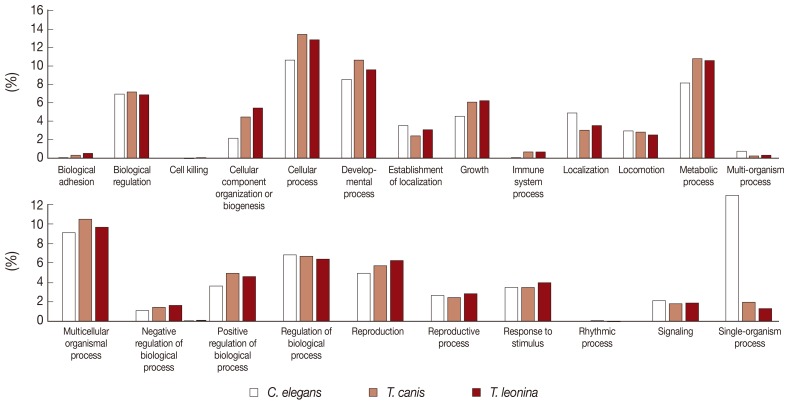
Fig. 3Comparative analysis of Molecular Function Level 2 GO terms associated with C. elegans, T. canis, and T. leonina enriched UniScripts. Molecular Function Level 2 GO terms were derived for stage-enriched UniScripts using CLC Genomics Workbench. A major dichotomy was evident in the binding and catalytic activity subcategories.
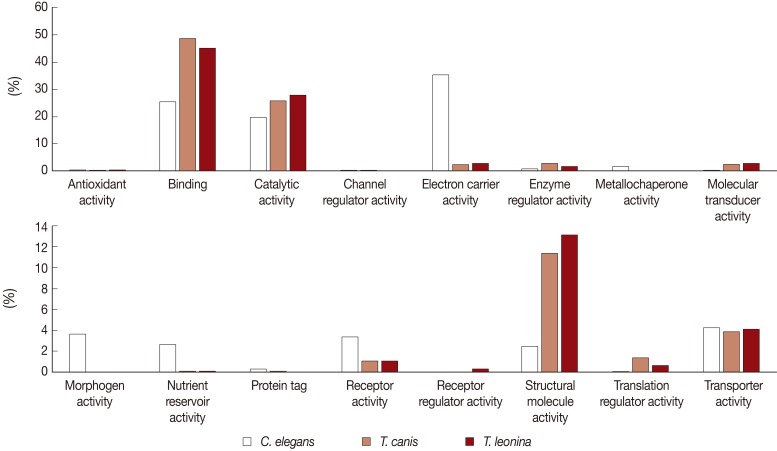
Table 1.Statistics of EST of Toxascaris leonina and Toxocara canis
Table 1.
|
T. leonina
|
T. canis
|
|
Total clones sequenced |
2,880 |
7,949 |
|
ESTs submitted for BLAST search |
585 |
2,070 |
|
Contigs |
170 |
808 |
|
Singletons |
415 |
1,262 |
|
No. of EST clusters |
42 |
117 |



