Abstract
Cystic echinococcosis (CE) is a zoonotic infection caused by Echinococcus granulosus larvae. It seriously affects the development of animal husbandry and endangers human health. Due to a poor understanding of the cystic fluid formation pathway, there is currently a lack of innovative methods for the prevention and treatment of CE. In this study, the protoscoleces (PSCs) in the encystation process were analyzed by high-throughput RNA sequencing. A total of 32,401 transcripts and 14,903 cDNAs revealed numbers of new genes and transcripts, stage-specific genes, and differently expressed genes. Genes encoding proteins involved in signaling pathways, such as putative G-protein coupled receptor, tyrosine kinases, and serine/threonine protein kinase, were predominantly up-regulated during the encystation process. Antioxidant enzymes included cytochrome c oxidase, thioredoxin glutathione, and glutathione peroxidase were a high expression level. Intriguingly, KEGG enrichment suggested that differentially up-regulated genes involved in the vasopressin-regulated water reabsorption metabolic pathway may play important roles in the transport of proteins, carbohydrates, and other substances. These results provide valuable information on the mechanism of cystic fluid production during the encystation process, and provide a basis for further studies on the molecular mechanisms of growth and development of PSCs.
-
Key words: Echinococcus granulosus, encystation process, differentially expressed gene, protoscolex, transcriptome
INTRODUCTION
Cystic echinococcosis (CE), also known as hydatid disease, is a chronic neglected zoonotic disease caused by the larvae of
Echinococcus granulosus that endangers human health and leads to huge economic losses in animal husbandry [
1]. Recent epidemiological studies have shown that at least 270 million people (58% of the total population) are at risk of CE in Central Asia, including Mongolia, Kazakhstan, Kyrgyzstan, Tajikistan, Turkmenistan, Uzbekistan, Iran, Pakistan, and western China [
2,
3]. The larval stage (hydatid cyst) of
E. granulosus is characterized by long-term growth in the internal organs of humans and other intermediate hosts, especially in the liver and lungs [
4]. Benzimidazoles have been widely prescribed to treat CE, despite its main disadvantages of poor absorption and high hepatotoxicity [
5,
6]. Thus, the need to develop new chemotherapeutics against CE should be emphasized
The hydatid cyst is composed of cyst wall, cystic fluid, protoscoleces (PSCs), and brood capsules. The cyst wall mainly consists of an outer protective acellular mucin-rich laminated layer [
7] and an inner layer of germinal cells (blastoderm) [
8]. The germinal layer has many cellular nuclei which can produce groups of vesicles into the cyst lumen. These vesicles become brood cysts after cell vacuolation and then develop into the initial PSCs [
9]. Secondary hydatid cysts are formed by encystation during both
in vivo development and
in vitro culture of the PSCs [
10]. However, despite the fact that cyst volume and the amount of cystic fluid gradually increases during the process of encystation, as well as the fact that the echinococcus cyst fluid is the internal environment for the growth and development of germinal cells and PSCs, the functional genes and the key metabolic pathways responsible for the encystation process remain unknown.
Recently, second-generation high-throughput RNA sequencing technology has been used to analyze the expression level of the whole genes or transcriptomes in a sample, under specific physiological conditions. As such, it plays an important role in characterizing gene expression and regulation [
11]. Whole-genome sequencing, proteomic analyses, and transcriptomic investigations have been used to identify the differentially expressed genes (DEGs) and proteins in the different life stages (adult, oncosphere, hydatid cyst, larval worms, and pepsin/H+-activated PSCs) of
E. granulosus [
12–
15]. However, the transcriptomic features of PSCs in the encystation process have not yet been described. The aim of this study was to evaluate the mechanism of cyst fluid formation in PSCs by RNA-seq and to determine the key DEGs and the main metabolic pathways used by PSCs to produce the energy needed to maintain their growth and development during the encystation process.
MATERIALS AND METHODS
Protoscolex collection and RNA extraction
Echinococcus granulosus hydatid cysts were collected from the liver of a naturally infected sheep obtained from a slaughterhouse in Urumqi, Xinjiang, China. The hydatid fluid was aspirated aseptically using a 50 ml syringe. The PSCs were then collected from the hydatid fluid under aseptic conditions and washed 15 times with sterile PBS. Afterward, the brood capsules were digested in 1% (w/v) pepsin for 30 min at 37°C, to release the PSCs. During digestion, the samples were agitated every 5 min and observed under an inverted microscope until the tissues were completely digested. The collected PSCs were washed with PBS supplemented with penicillin and streptomycin (Sigma, St. Louis, Missouri, USA), and stained with 0.4% Trypan blue. Batches with a viability rate of over 90% were selected for further analysis. The PSC culture conditions were previously described by Liu et al., 2018 [
16]. The PSCs were cultured in RPMI 1640 medium (Gibco, Grand Island, New York, USA) (pH 7.2), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The average number of PSCs in the monophasic culture medium was 4,000 per bottle, and the incubator conditions were 37°C and 5% CO
2. The culture medium was changed every 2 to 3 days. Different stages of culture medium were isolated based on the morphological classification [
17]. The isolated PSCs at different stages were stored in liquid nitrogen for RNA extraction. PSC tissues were collected from 15 batches of 5 culture periods (3 replicates each): EgPSC_0, 10, 20, 40, and 80 days. Total RNA was extracted from all samples using a TRIZOL-Reagent kit (Invitrogen, Carlsbad, California, USA), according to the manufacturer’s instructions. RNA concentration and integrity were detected by ultraviolet spectrophotometry and agarose gel electrophoresis.
Construction of strand-specific cDNA libraries and sequencing
A strand-specific cDNA library was constructed on Oligo (dT) bead-enriched PSC mRNA using an Illumina® TruSeq® RNA Sample Preparation Kit v2 (Illumina, San Diego, California, USA), according to the manufacturer’s instructions. The cDNA libraries were combined according to the effective concentration and target data volume, and sequenced by high-throughput sequencing using Illumina His seq X (Illumina, California, USA) (Majorbio, Shanghai, China).
Assembly and annotation
Analysis on inter-sample correlation
RNA-Seq by expectation-maximization (RSEM) (
http://deweylab.github.io/RSEM/) quantitatively analyzes the expression levels of genes and transcripts and can be used to analyze the DEGs between different samples in order to determine the regulation mechanism of genes by combining functional information [
20]. In addition, RSEM distinguishes transcript subtypes of the same genes using a maximum likelihood abundance estimation model [
21]. A Venn diagram analysis was performed to display genes that were specific and co-expressed between samples.
Since there were three biological replicates per period, raw data were statistically analyzed directly using the DESeq2 software, which is based on the negative binomial distribution [
22]. EgPSC_0d was used as the control. Differentially expressed genes (DEGs) showing threshold settings (
P-adjust<0.05 & |log2FC1|>=1), transcript expression folds greater than 2 per group, and
P-values under 0.05 after multiple-test corrections were selected [
23].
Twelve representative DEGs (
P-adjust<0.05 & |log2FC|>=1) were randomly selected for validation using Illumina His seq X (Illumina, California, USA) sequencing data, including 6 up-regulated DEGs and 6 down-regulated DEGs [
24]. Primer 5.0 and Oligo 7.0 software were used to design and evaluate specific primers (
Table 1). Actin II was employed as an endogenous reference gene based on previous publications [
25] and these primers were synthesized by Tsingke Biotech Company (Beijing, China). All 13 RNA templates were reverse-transcribed to cDNA by using the Prime ScriptTM RT reagent Kit (Takara, Tokyo, Japan), according to the manufacturer’s instructions. The cDNAs were diluted 10 times with nuclease-free water. Qrt-PCR was performed using a SYBR Premix Ex Taq GC kit (Takara) with a reaction mix comprised of 10 μl of SYBR Q-PCR mix (2×), forward and reverse primers (0.8 μl, 10 μM each), and 4 μl of diluted cDNA. Double-distilled H
2O was added to bring the volume to 20 μl. The thermal cycles were performed as follows: 95°C for 2 min and 40 cycles of 95°C for 10 sec, 55°C for 10 sec and 72°C for 20 sec. The dissolution curve was plotted at 65°C for 5 sec, 95°C for 5 sec and 4°C for 30 sec. Each DEG was assayed in triplicate using a CFX-96 TouchTM Real-Time PCR Detection System. Transcript level of the DEGs were calculated using the 2
−ΔΔCt method [
26]. Inter-sample statistical analysis was performed using T-test.
RESULTS
Protoscoleces and total RNA
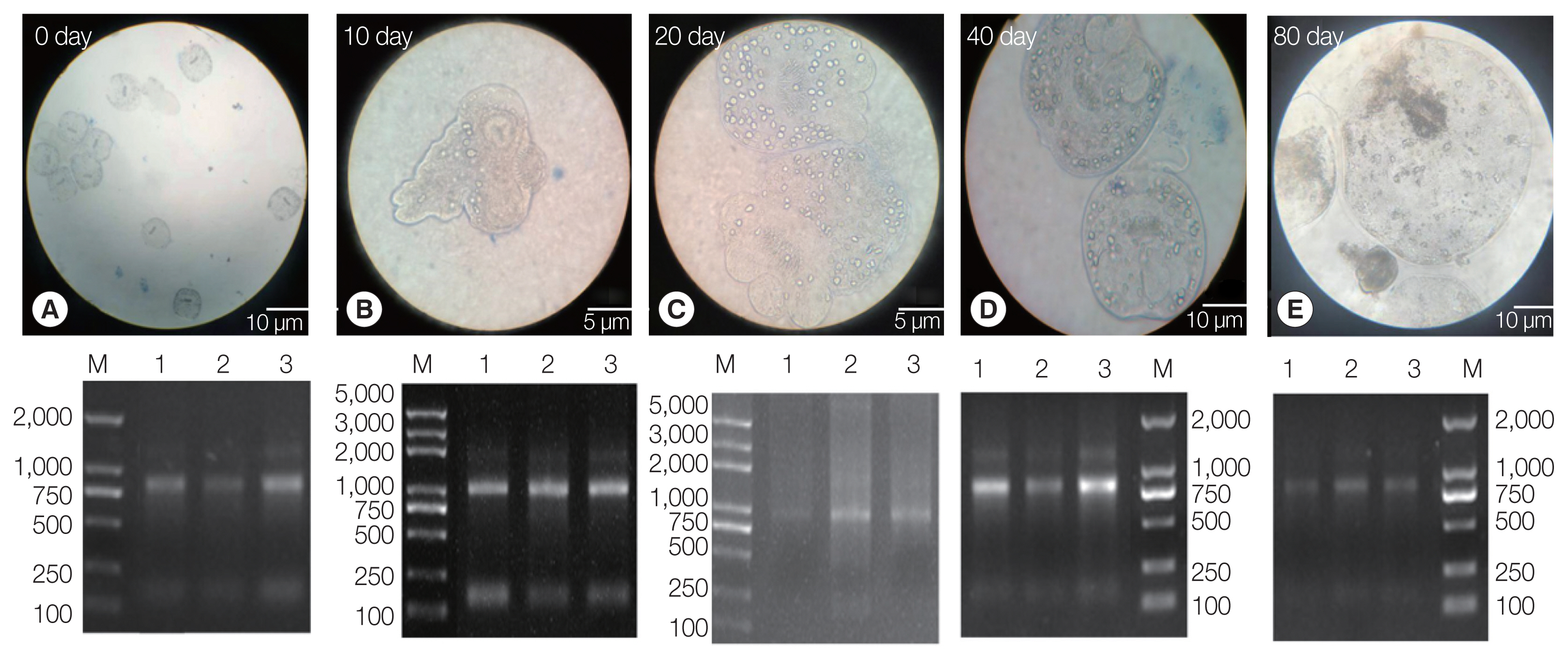
The PSCs were invaginated at the beginning of the
in vitro culture. Calcareous corpuscles and apical hooks were observed under an inverted microscope (
Fig. 1A). After 10 days of incubation, the morphology of the PSCs was found to vary greatly, with the apical hook and sucker becoming evaginated (
Fig. 1B). After 20 days, microcysts appeared with a thin laminated layer and the apical hooks on the scolex moved toward the central position of these micro-cysts. Moreover, the calcareous corpuscles of the micro-cysts were clearly visible (
Fig. 1C). After 40 days, microcysts with a laminated layer were clearly observed, and the apical hooks and suckers were not completely degraded (
Fig. 1D). On the 80th day, the apical hook and sucker disappeared or degraded and the calcareous corpuscles decreased, while the size of the cysts increased (
Fig. 1E).
Three batches of the PSCs were collected at 5 culture periods (EgPSC_0d, EgPSC_10d, EgPSC_20d, EgPSC_40d, and EgPSC_80d). Total RNA on the gel showed slightly degraded RNA bands.
Sequence assembly and annotation
The cDNA library of each incubation period generated an average of 51,300,044 clean reads with a total of 7,597,846,677 nucleotides in each sample (
Table 2). After alignment and assembly, 14,903 genes and 32,401 transcripts were obtained. Among these, 3,584 new genes and 21,082 new transcripts were found that had not been previously reported in the parasite. Among 32,401 transcripts, 20,511 were annotated by GO (63.3%), 13,730 by KEGG (42.4%), 17,152 by COG (52.9%), 28,188 by NR (87%), 16,511 by Swiss-Prot (51.1%), and 17,874 by Pfam (55.2%). Additionally, the Venn diagram analyzed 20,511 functionally annotated transcripts. A total of 4,458 reference transcripts and 53 new transcripts were simultaneously compared to the above databases and corresponding annotations were obtained. The transcripts length ranged widely, although the greatest proportion of these (34.7%) were longer than 1,800 bp. Transcripts shorter than 200 bp were the least common (1.3%). The resulting transcriptome raw reads dataset was submitted to the NCBI Short Read Archive (SRA) database under accession number SRP172517.
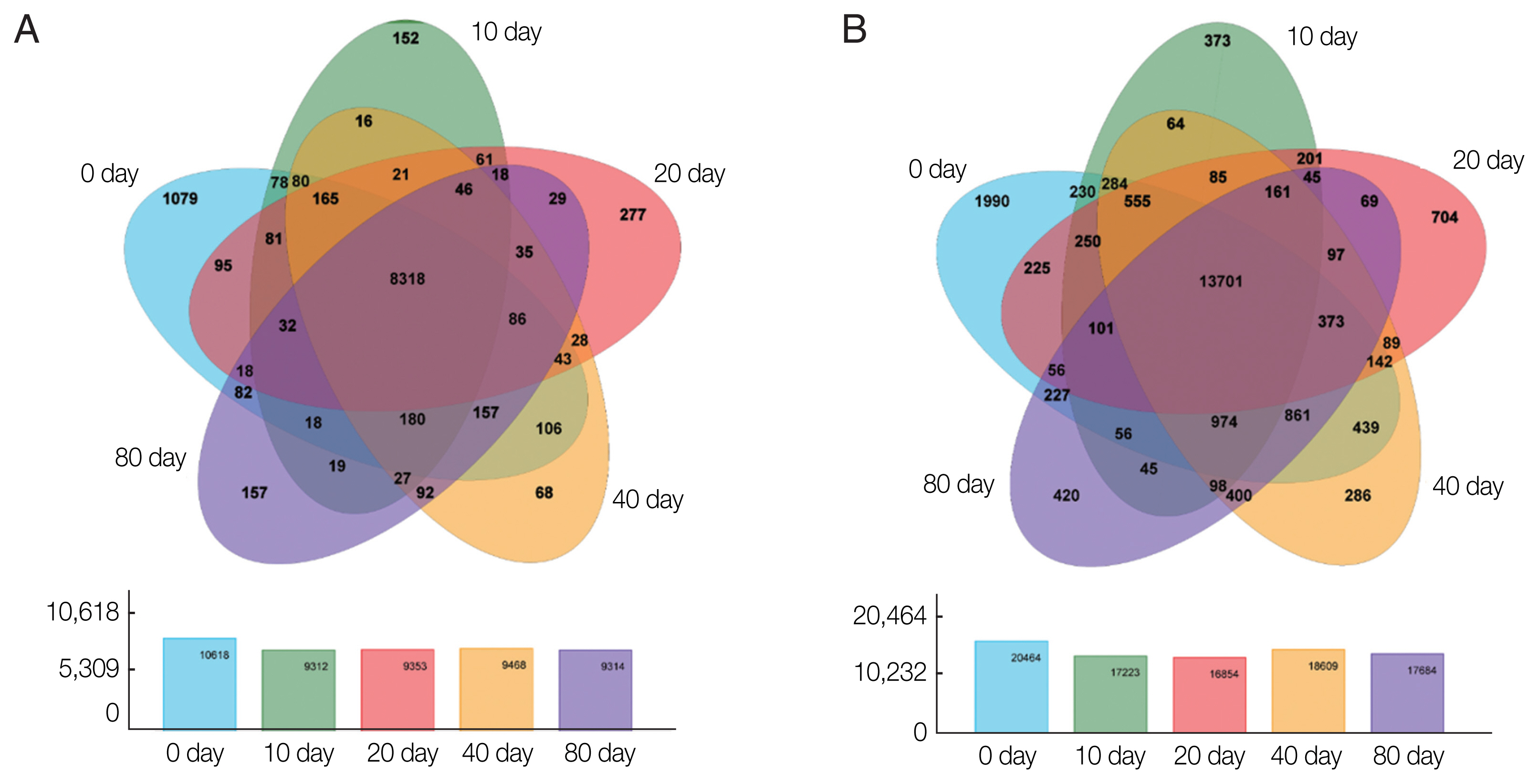
Based on the expression matrix, an inter-sample Venn diagram analysis was performed to obtain the co-expressed and differentially expressed genes (
Fig. 2A) or transcripts (
Fig. 2B) between the incubation period groups. Intriguingly, although 14,903 genes and 32,401 transcripts were obtained by transcriptome sequencing at the different developmental stages of PSCs in the encystation process, these genes and transcripts were not always presented in every period of the cyst formation process. Conversely, various genes and transcripts were specifically expressed in different stages. At EgPSC_0d, the number of stage-specific genes and transcripts was largest, at 1079 and 1990, respectively, while the stage-specific genes and transcripts at the 20d stage were 277 and 704, respectively.
A total of 1,991 up-regulated and 2,517 down-regulated DEGs were obtained through parameter setting. A set of genes composed of 1,991 up-regulated DEGs was established to determine a functional classification compared with the COG database. Results showed that 25% of these genes were classified to the poorly characterized category which described an unknown function. Moreover, 7.2% of these genes were classified as cellular processes and signaling, which described as intracellular trafficking, secretion, and vesicular transport (
Supplementary Table S1).
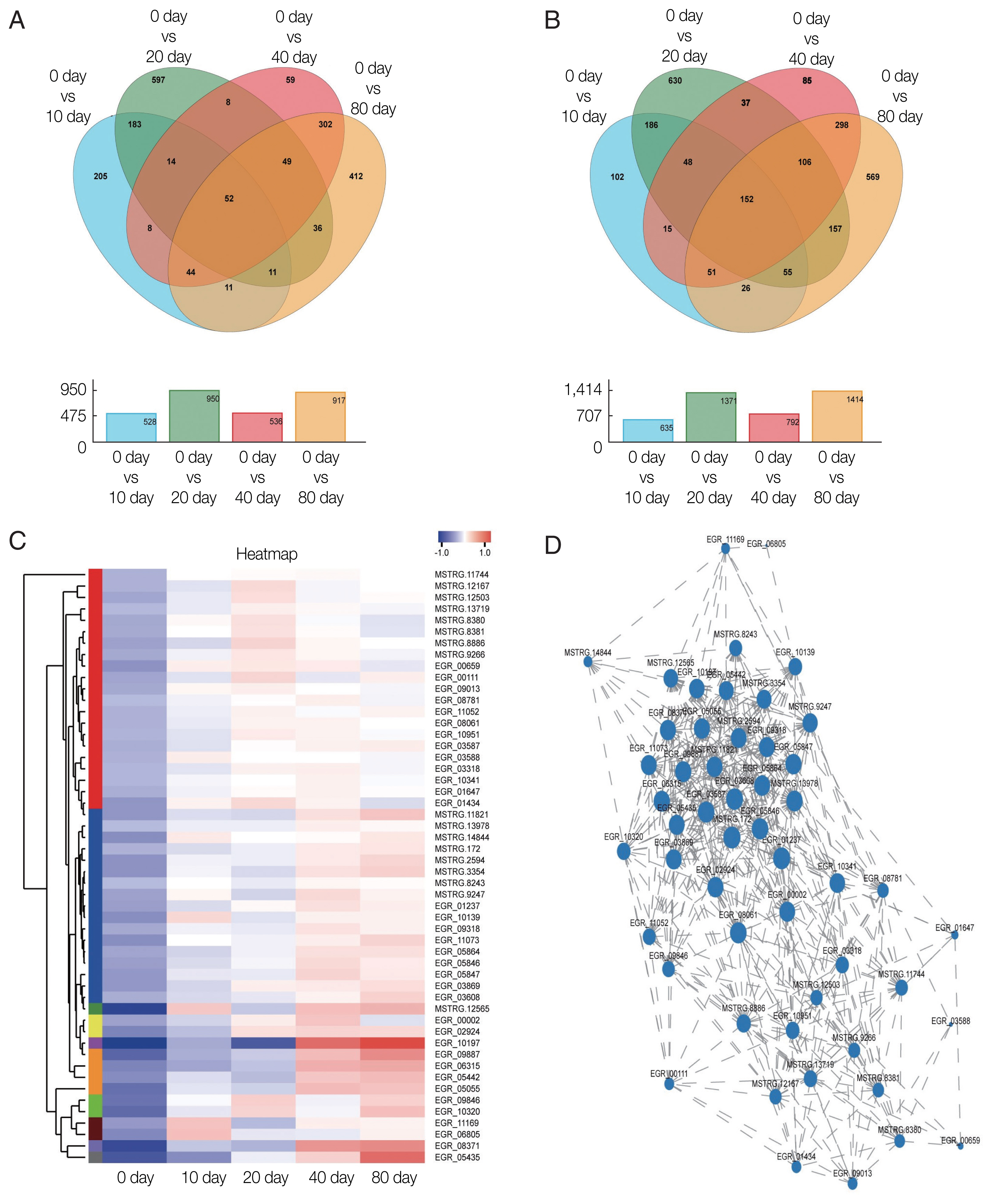
The Venn diagram analysis revealed that 1,991 DEGs were up-regulated and 2,517 were down-regulated. The expression of 52 DEGs consistently increased during encystation (
Fig. 3A), while 152 genes consistently declined (
Fig. 3B). A cluster heat map analysis of 52 consistently up-regulated DEGs (
Fig. 3C) showed that the hypothetical proteins, EGR_10197 and EGR_05435, increased significantly during late encystation. Gene1117 (EGR_10197) is an integral component of the membrane and is classified into the cellular component term of the GO database. Of the 52 DEGs, 13 genes were annotated to the COG database and were found to serve numerous important biological functions, including amino acid transport and metabolism, intracellular trafficking, secretion, and vesicular transport (
Table 3). After analyzing the correlation coefficients between the 52 DEGs with FDR <0.05 (
Fig. 3D), a visualization network was obtained, indicating that cornifelin (EGR_03608), platelet glycoprotein (EGR_09887), and potassium voltage-gated channel subfamily C member 3 (EGR_05864) among others, were related to the expression of various other genes.
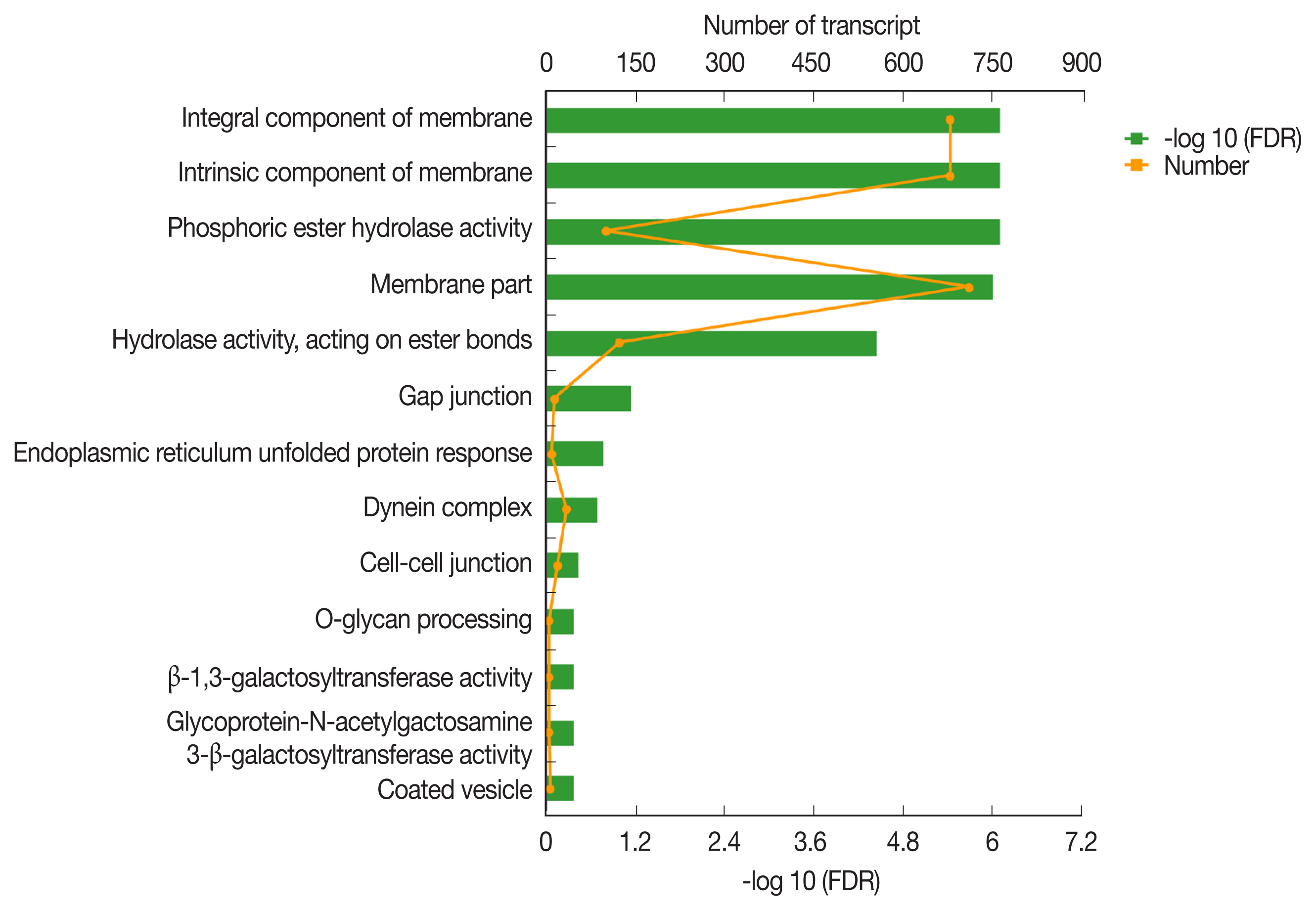
The GO classification of function-enriched 1,991 DEGs showed that 1,094, 148, and 263 DEGs were assigned to the cellular component (CC), biological process (BP), and molecular function (MF) categories, respectively, of which 709, 0, and 122 genes were significantly differentially expressed (FDR value <0.05). The top 3 predominant terms for BP were endoplasmic reticulum unfolded protein response, O-glycan processing, and ion transport. For CC, they were integral components of membrane, intrinsic components of membrane, and membrane part. For MF, they were phosphoric ester hydrolase activity, hydrolase activity, acting on ester bonds, and beta-1,3-galactosyltransferase activity (
Fig. 4).
KEGG pathway enrichment analysis revealed that 1,991 DEGs were classified into 291 pathways. The most significantly enriched pathways was the vasopressin-regulated water reabsorption metabolic pathway (FDR<0.05) (
Fig. 5), followed by the pantothenate and CoA biosynthesis (map00770) and Hippo signaling pathway (map04391). The morphological change of the PSCs was greatest in the first 10 days. We therefore performed GO and KEGG enrichment chord analysis of the top 10 GO terms and KEGG pathways with an FDR value <0.05 on the DEGs between EgPSC_0d and EgPSC_10d. The DEGs of EgPSC_0d and EgPSC_10d were 43 and 61, respectively (
Supplementary Figs. S1,
S2).
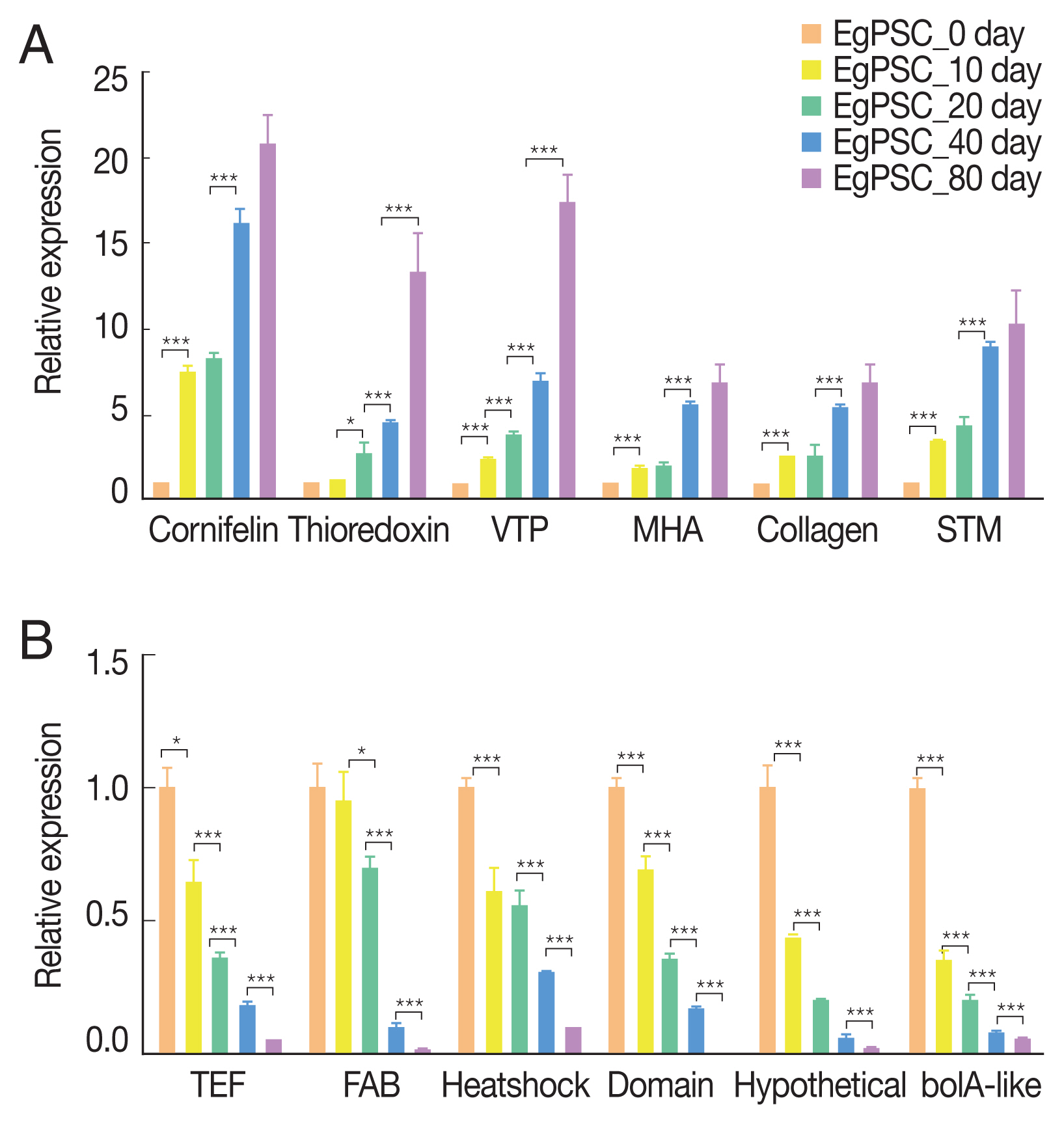
Six up-regulated and 6 down-regulated DEGs were randomly selected to validate the high-throughput RNA sequencing data and DEGs by Qrt-PCR. The Qrt-PCR result of 6 up-regulated DEGs: cornifelin, thioredoxin-related trans-membrane protein, vesicle transport protein, minor histocompatibility antigen, collagen and signal transducing adapter molecule (
Fig. 6A). While the 6 down-reg-ulated DEGs: transcription elongation factor, fatty acid-binding protein, heat shock protein, LIM and SH3 domain protein, hypothetical protein and bolA-like protein (
Fig. 6B). The expression of these genes in PSCs was validated by Qrt-PCR, which was highly correlated with the results obtained by RNA-Seq.
DISCUSSION
The parasite
E. granulosus is a cestode tapeworm that acts as a causative agent of CE, one of the 17 previously neglected tropical diseases recently prioritized by the World Health Organization (WHO) [
15]. Hydatid cysts develop into the pre-adult form during its life-cycle and can either differentiate into an adult worm (strobilation) in the terminal host (dogs), or into a secondary hydatid cyst in the intermediate host (humans and livestock) [
27]. Furthermore, cysts can develop in almost any internal organ or tissue through hematogenous dissemination, including the heart, bones, and nervous system [
28]. Early infection is asymptomatic. However, increases in cyst volume can lead to physical damage due to the mechanical compression of the surrounding tissues and organs [
29]. The major causes of morbidity and mortality include pressure exerted by the cysts, cyst location in a sensitive organ, or cyst rupture caused by a spontaneous or external force (traumas or surgery), resulting in anaphylaxis or dissemination of the secondary infection [
30]. Thus the need of novel prevention and control strategies based on basic and applied
E. granulosus biology, especially genomic, transcriptomic, and proteomic analyses should be emphasized. Our understanding of key DEGs and the main metabolic pathways of PSCs in the encystation process remains limited.
Previous investigations found that PSC encystation generally appeared during the first week of in vitro incubation, in which micro-cysts with a complete laminated layer were observed on day 20. Fully developed micro-cysts were observed between days 38 and 42 [
31]. Corresponding nodes were observed in our laboratory at 10 d, 20 d, and 40 d, respectively, and cysts were large enough following 80 days of in vitro culturing. These phenomena varied slightly from those reported in previous studies, which may be attributed to the use of different culture media and genotypes for bladder formation, and slightly different development processes [
17].
We obtained 32,401 transcripts and 14,903 genes and demonstrated that 1,991 and 2,517 genes were significantly up- and down-regulated, respectively. The GC content is an important characteristics of the genome base sequence, which reflects the structure, function, and evolutionary information of the genes [
32]. The average GC content of the
E. granulosus transcriptome (46.4%) was similar for both its genome (42.1%) and coding regions (49.3%) [
33]. Early studies estimated the GC contents of
S. mekongi,
B. malayi, and
S. mansoni had to be 34.1%, 30.5%, and 35.3%, respectively [
24,
34]. The GC content of
E. granulosus was higher, compared to other parasite species. Furthermore, some genes or transcripts could not be annotated and were defined as hypothetical proteins [
35].
Gene-encoding proteins involved in several signaling pathways, like putative G-protein coupled receptor (GPCR), tyrosine kinases, and serine/threonine protein kinase were predominantly up-regulated during the encystation process of PSCs. These signaling pathways play major roles in key functions, including movement, development, and reproduction in parasites [
36]. Our findings are consistent with those of previous studies on
S. mekongi [
24] and
S. japonicum [
37]. A previous study classified over 60 putative GPCRs of
E. granulosus as potential anthelminthic drug targets [
14]. We filtered out coding putative GPCR genes with low expression and non-expression using transcript sequencing, narrowed down to 6 genes. Interestingly, EGR_00873 was significantly up-regulated from EgPSC_0d to EgPSC_20d. Tyrosine kinase and serine/threonine protein kinase did not increase during the first 10 days of PSCs
in vitro culture, but were significantly up-regulated in the later stage. The members of these pathways characterized here may represent novel drug targets for anti-parasitic intervention. However, further studies are necessary to confirm our results.
The majority of invertebrate species have shown an immune response that can inactivate and eliminate penetrating parasites, especially those involving the formation of reactive oxygen species (ROS) [
38]. ROS is a key trigger of the inflammatory activation of macrophages in malaria and can damage proteins, carbohydrates, and DNA, which are detrimental to parasites [
39]. Protective antioxidant proteins are produced by the parasites to neutralize the host’s immune response and reduce the oxidative damage caused by oxygen free radicals [
40]. In this study, we identified three major antioxidant proteins in PSCs, namely cytochrome c oxidase, thioredoxin glutathione, and glutathione peroxidase. Cytochrome c oxidase, an up-regulated DEGs, has the molecular function of binding to heme and transporting proteins in the mitochondrial electron transport chain, according to the COG database annotation. This is consistent with previous observations, wherein it was found to regulate the mitochondrial oxidative metabolism of
E. granulosus [
41]. Previous reports have suggested that cytochrome c oxidase dysfunction was associated with increased mitochondrial ROS production, as well as inflammation and apoptosis [
42,
43]. Considering external environmental pressures, cytochrome c oxidase in PSCs may play essential roles in the antioxidant defenses during the encystation process. The thioredoxin glutathione system is another well-characterized antioxidant defense which plays a critical role in maintaining the redox balance in
S. japonicum [
44],
E. granulosus [
12],
T. brucei [
45], and
S. mansoni [
40]. In this study, three types of proteins, namely thioredoxin, thioredoxin-related transmembrane proteins, and thioredoxin domain-containing proteins, were analyzed using differential expression analysis. Among these, glutathione peroxidase (EGR_06748) presented continuously high expression levels in PSCs during the encystation process, and is known to be involved in the clearance of cytotoxic and genotoxic compounds and protection against oxidative damage [
46]. Studies have previously reported that glutathione peroxidase in
E. granulosus has the ability to bind non-substrate molecules, particularly anthelmintic drugs [
47], and acts as an essential enzyme for the survival of schistosomiasis in the redox environment. It has therefore been actively investigated as a potential drug target [
48].
We analyzed the key genes identified by genomic [
33], proteomic [
49], and transcriptomic [
12] studies of
E. granulosus, and found that the expression levels of certain genes changed significantly during the process of PSC encystation. For example, in heat shock proteins, the gene EGR_09650 was only expressed in adult worms, while EGR_10561 was highly expressed in the hydatid cyst membrane [
33]. However, we found that both EGR_09650 and EGR_10561 were expressed in PSCs. It is worth noting that EGR_10561 consistently showed the highest expression among all genes coding for heat shock proteins during the encystation process of PSCs.
In terms of glycosylphosphatidylinositol (GPI)-anchored wall transfer protein, an important protein exclusive to
E. granulosus, showed a low expression level of the encoding gene, EGR_06221, during PSC encystation. This could be attributed to the fact that heterologous proteins are covalently anchored to the PSC cell wall by fusing with the anchoring domain of GPI-anchored cell wall transfer protein [
50]. However, PSCs were cultured in vitro and there was no heterogeneous source protein in this study. Tetraspanins are transmembrane proteins previously described as potential vaccine candidates for other helminth infections and are also found in the membranes of the tegument and extracellular vesicles of
O. viverrini [
51]. Their function is associated with virulence and pathogenesis in some organism [
52]. A wide variety of molecules have been found to be released by
E. granulosus, including tetraspanin proteins, channel proteins that transport lipids, and nucleic acids [
53,
54]. We identified 2 tetraspanin-encoding genes (EGR_11042 and EGR_06311) that showed strikingly up-regulate trends during the encystation of PSCs. We speculated that the overexpression of genes related to protein transport in PSCs may be a compensatory adaptation mechanism to the external environment to satisfy their own encystation processes.
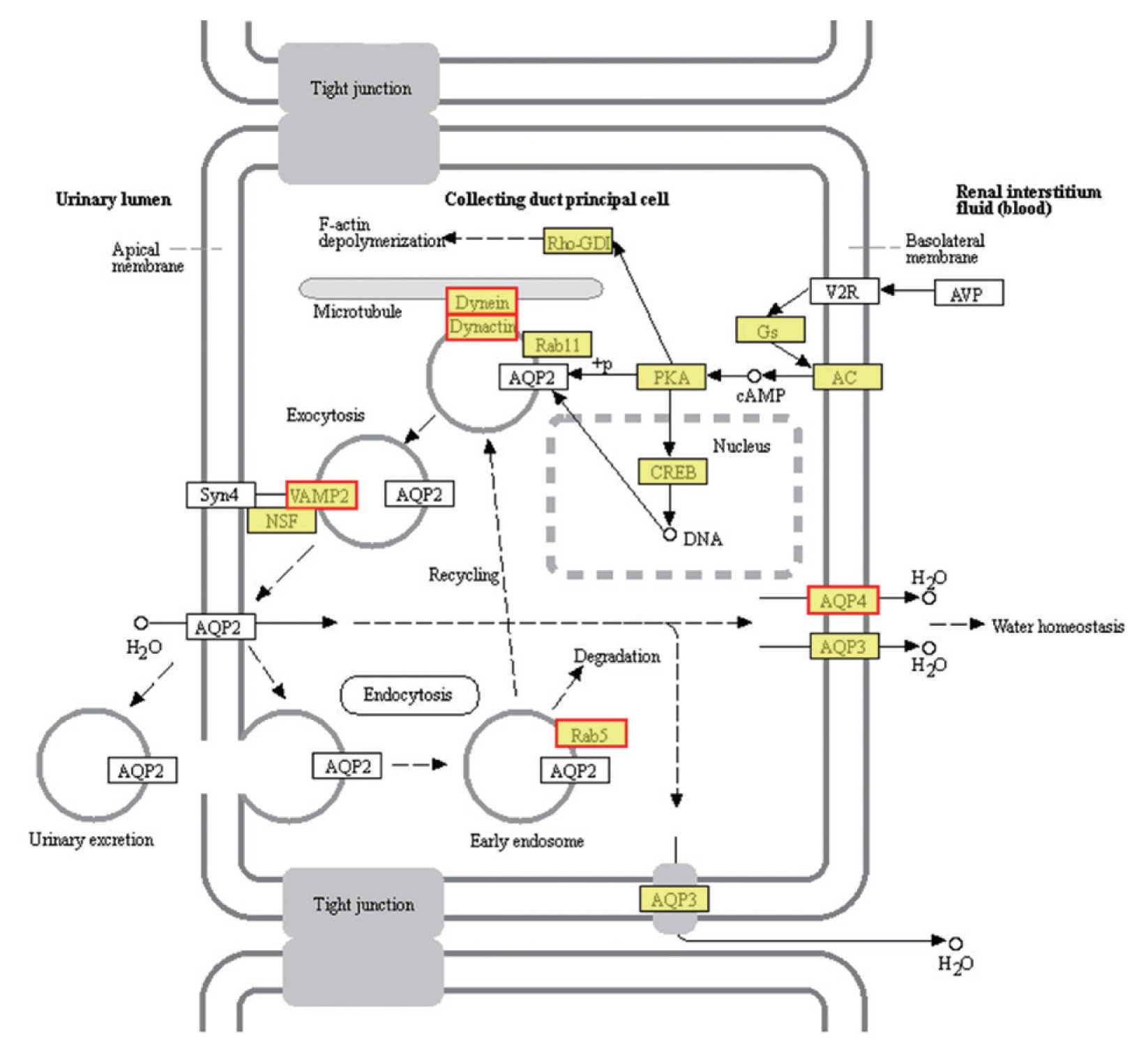
The most enriched KEGG pathways associated with the 1,991 up-regulated DEGs of PSCs during the encystation process were the vasopressin-regulated water reabsorption metabolic pathways (map04962), which were associated with a total of 24 DEGs (
Supplementary Table S2). Dynein light chain (DLC), ras-related protein, synaptobrevin-like protein, and aquaporin 4 (AQP 4) were also up-regulated during the PSCs encystation process. Vasopressin-regulated water reabsorption metabolic pathways have previously been reported to play a critical role in regulating water, urea, and sodium transport, and are necessary to maintain the water balance in the body [
55]. However, due to the special structure and life history of the parasite, there is no aquaporin 2, vasopressin, or the corresponding V2 receptor (V2R) genes in PSCs. Previous reports have indicated that DLC is involved in the transport of proteins, carbohydrates, and other substances, as well as in retrograde vesicles [
56]. Ras-related proteins are known to be involved in vesicle transport after activation. Parasites can secrete extracellular vesicles to achieve intercellular communication and transfer biologically active molecules to the host cells to modulate host immune response [
57]. Vesicle transport is therefore considered an important pathway for the transport of substances and the transmission of information between parasites and the external environment [
49]. In this study, we detected AQP4 in the cell membranes of PSCs. The eukaryotic parasites
S. japonicum [
58] and
S. mansoni [
59] permeate and regulate water reabsorption by activating AQPs. Therefore, studying the role of vasopressin-regulated water reabsorption metabolic pathways in PSCs can provide useful insights in developing of new drug targets.
Notes
-
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supplementary Information
Supplementary Table S1.
COG functional classification of all of the up-regulated differently expression genes
Supplementary Table S2.
The 24 incolved in vasopressin-regulated water reabsorption metabolic pathways (map 04962) DEGs
Supplementary Fig. S1.
GO enrichment chord analysis of the top 10 GO terms with an FDR value<0.05 on the DEGs between EgPSC_0d and EgPSC_10d.
Supplementary Fig. S2.
KEGG enrichment chord analysis of the top 10 KEGG pathways with an FDR value<0.05 on the DEGs between EgPSC_0d and EgPSC_10d.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 81672045). The authors would like to express their gratitude to the editor and anonymous reviewers for their helpful comments and constructive suggestions.
Fig. 1Protoscoleces were cultured in vitro at different developmental stages. (A) Day 0 of culture. (B) Complete gestation was observed on day 10. (C) Day 20 of culture. (D) Day 40 of culture. (E) On day 80 of culture. Lower panel: agarose gel electrophoresis images with total RNA extracted from the protoscoleces, corresponding to each stage in the upper panel.
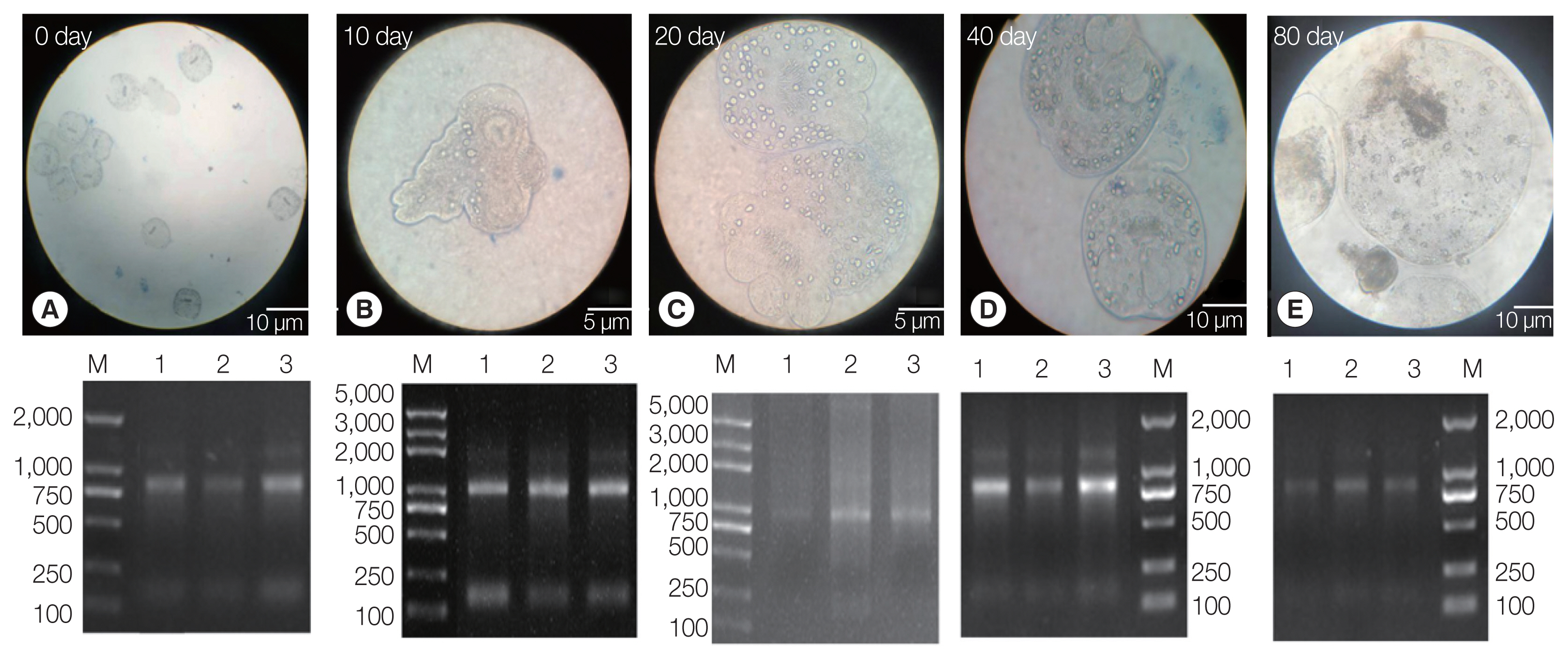
Fig. 2Venn diagrams showing differentially expressed genes among the experimental groups. Circles of different colors represent a set of (A) genes and (B) transcripts expressed in a sample, where the values represent the number of common and uniquely expressed genes/transcripts between a 2-or3-group sample. The sum of all numbers inside the circle represents the group. The sum of the number of expressed genes, the cross-region of the circle, represents the number of shared genes of each group of expressed genes/transcripts.
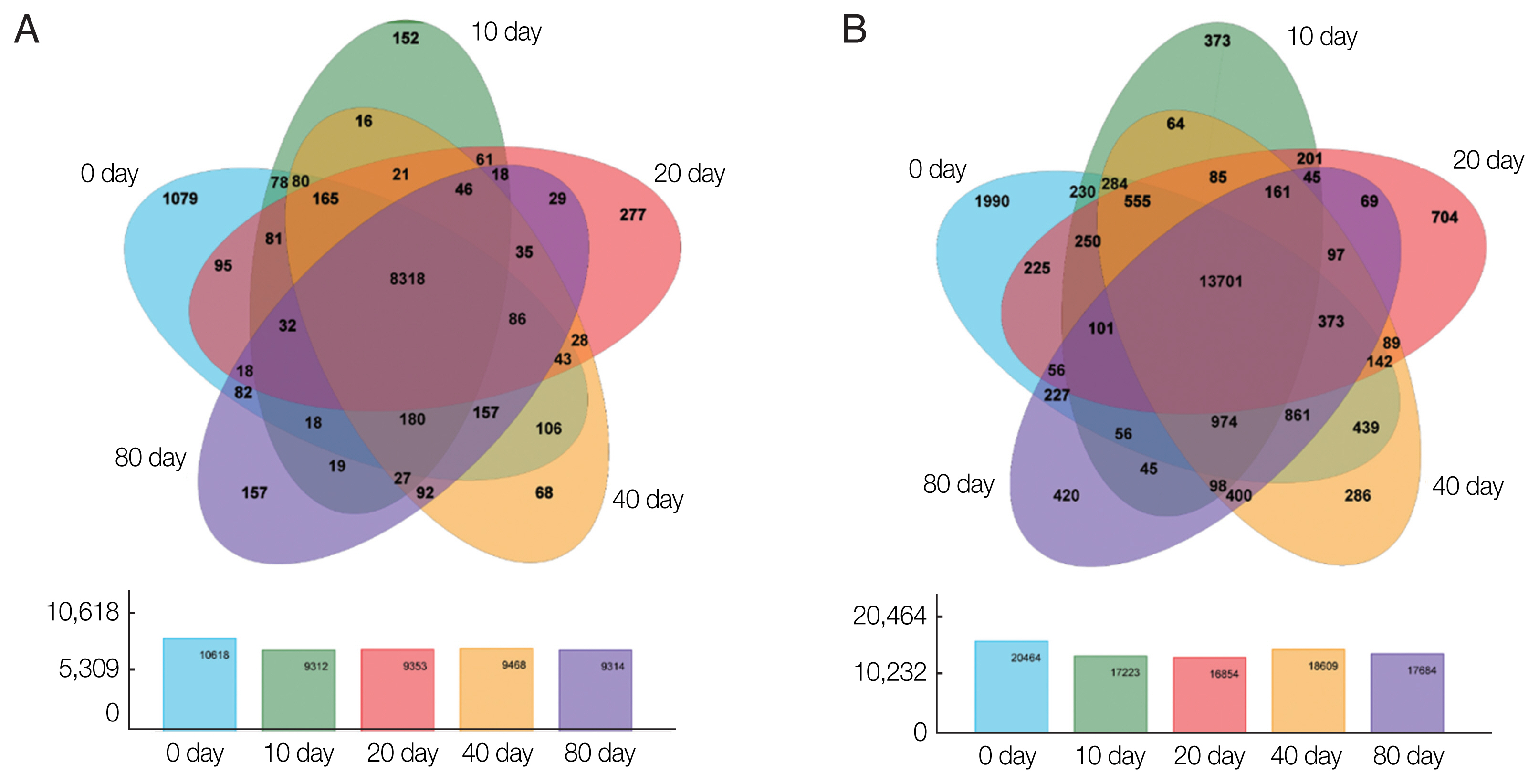
Fig. 3Transcription profiles of significantly differentially expressed genes. Venn diagram showing significantly (A) up-regulated DEGs and (B) down-regulated DEGs. (C) Hierarchical clustering map of 52 up-regulated DEGs showing overall TPM. log10 (TPM+1) values that are scaled and clustered. Red denotes up-regulated genes and blue denotes down-regulated genes. The upper clamps are sample clusters. The closer two sample branches are, the closer expression pattern of all genes in the two samples. (D) Correlation among 52 up-regulated DEGs. A node represents a gene. Lines between nodes represent a correlation between the levels of gene expression. The larger a node, the more a gene is correlated with other genes.
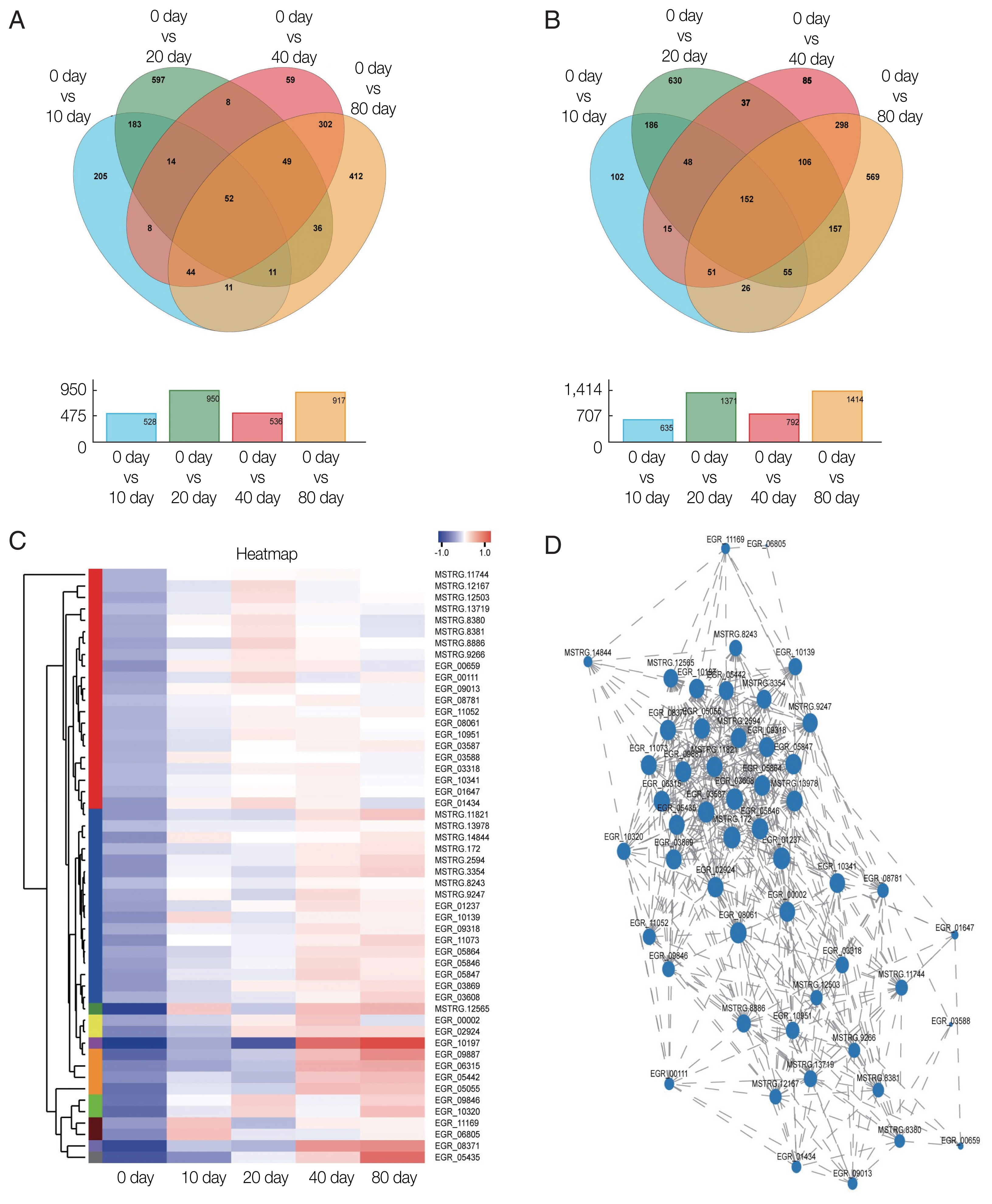
Fig. 4Gene ontology (GO) transcript classification. The broken line corresponds to genes at the different culture periods. The column indicates the enrichment level. The smaller the FDR −log10 (FDR) value, the more significant the GO term.
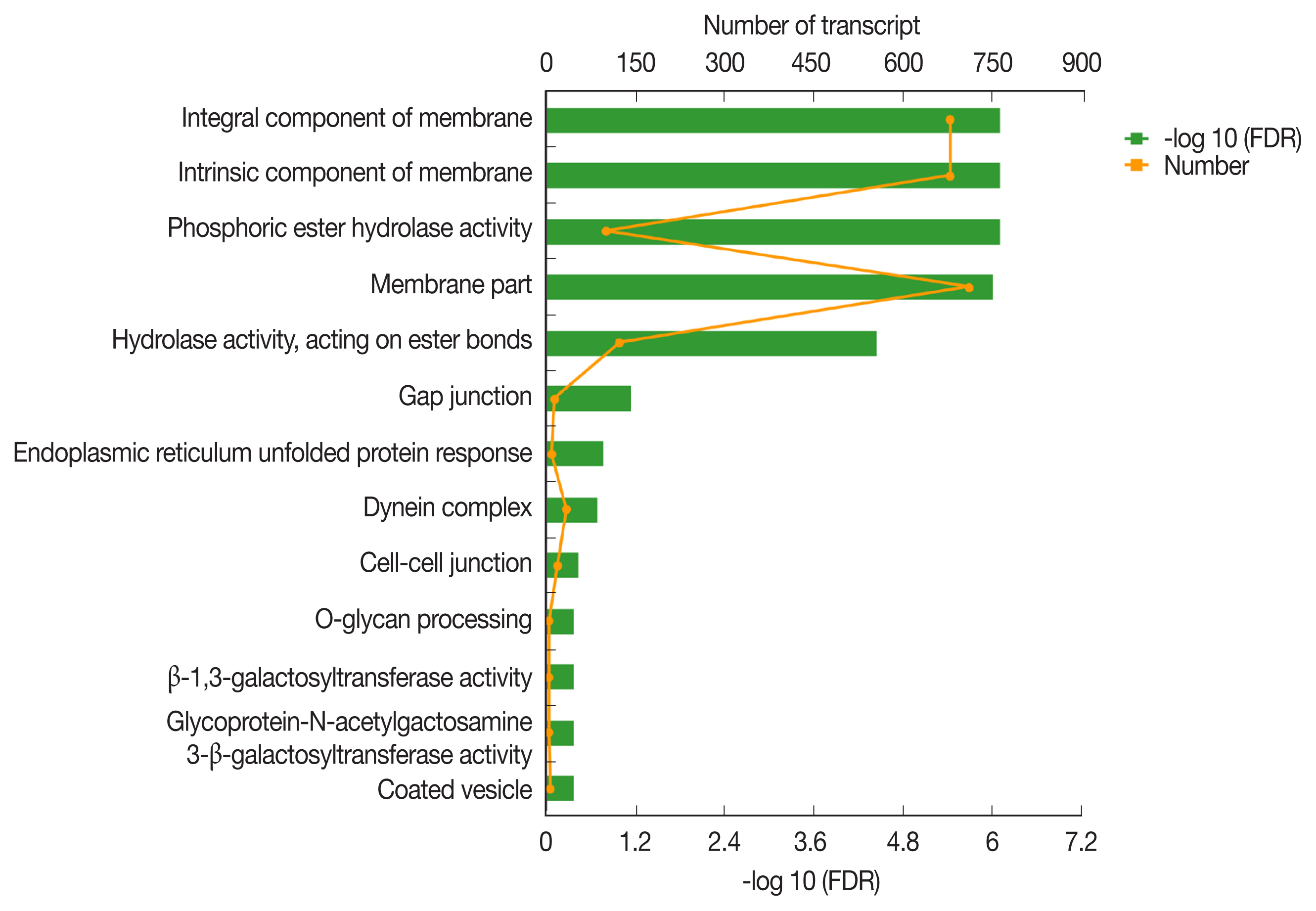
Fig. 5Vasopressin-regulated water reabsorption. Products with a colored background belong to the KEGG annotation results of this sequencing. A yellow background denotes the reference gene, and the red border indicates that the genes in the analyzed gene set are annotated.
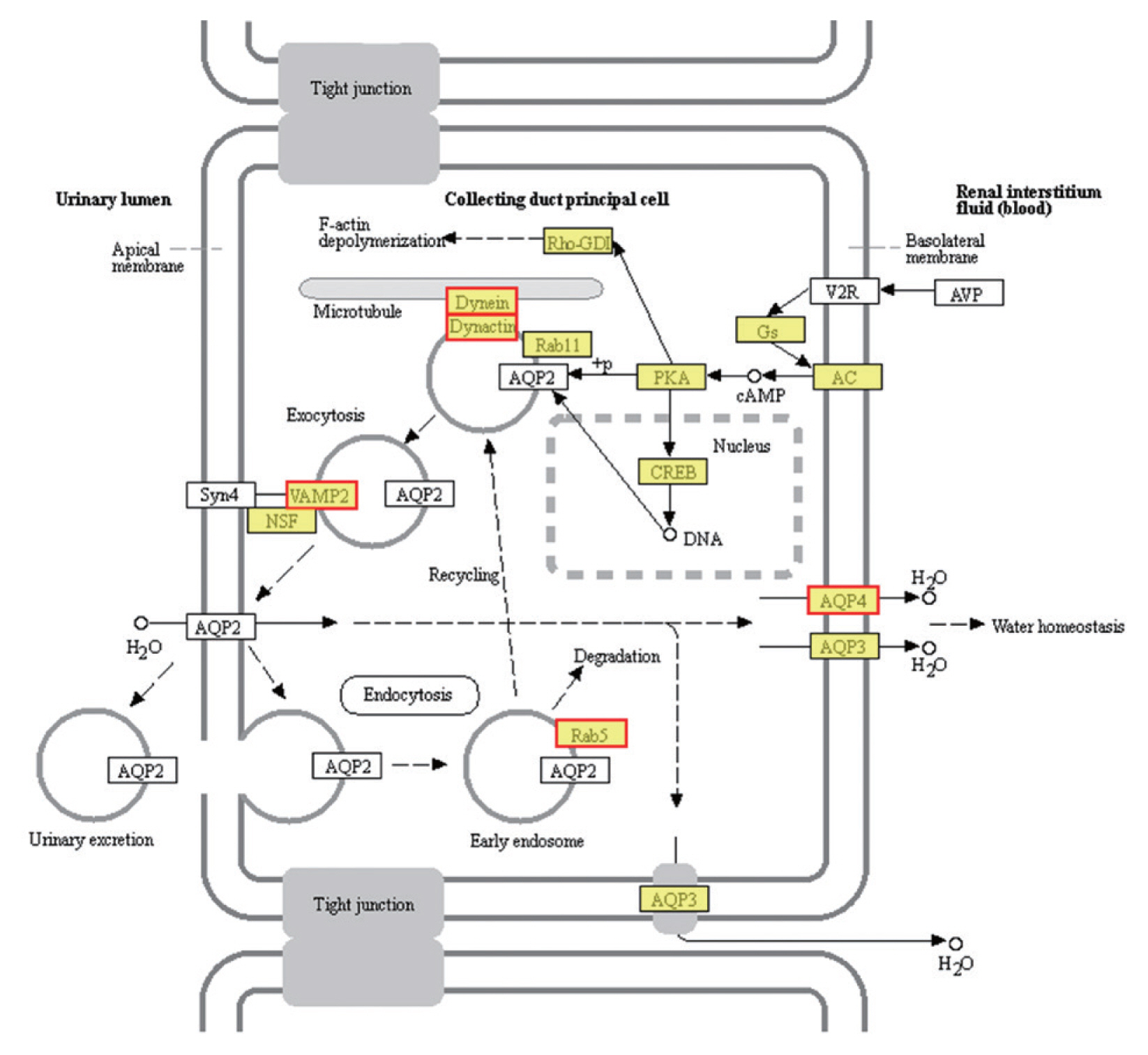
Fig. 6Verification of up-regulated (A) and down-regulated DEGs (B) by Qrt-PCR. Reference was actin II transcript. *P<0.01, **P<0.001, ***were considered extremely significant.

Table 1
Table 1
|
Gene name |
Annotation in gene database |
Forward (FP) and reverse primers (RP) |
Product size (bp) |
|
Up-regulated DEGs |
|
EGR_03608 |
Cornifelin |
FP: AGTCTGCTTCCATTGACGACCA |
149 |
|
|
RP: CCATCGCTGTCGCCTTCGGTA |
|
|
EGR_00384 |
Thioredoxin-related transmembrane protein |
FP: TGCAAATGTACGCTACTACCC |
88 |
|
|
RP: ACGTAAAGTAGGCTCAAGCAA |
|
|
EGR_01086 |
Vesicle transport through interaction with t-SNAREs 1A |
FP: AGATAGCAGCAACTACCCGTTT |
91 |
|
|
RP: TTCACATCAGCAGGCATGTCC |
|
|
EGR_06269 |
Minor histocompatibility H13 |
FP: TATTACCGTTTTCCGTCGAGA |
96 |
|
|
RP: GCGGCAAATATCTTAAACACC |
|
|
EGR_09743 |
Collagen, type XV, alpha |
FP: AAAGGACTCTACTGGACCTCG |
84 |
|
|
RP: GAAGTCCACTGATCGCACGTT |
|
|
EGR_08242 |
Signal transducing adapter molecule |
FP: TGATGCCCAAAATGCTACACC |
140 |
|
|
RP: CGAAATCGAATAGAGCACGAAC |
|
|
|
Down-regulated DEGs |
|
EGR_10426 |
Transcription elongation factor |
FP: TGATTGAGCCACTACCGAAGCA |
123 |
|
|
RP: AGTCCTCCAGGCATATTCCAC |
|
|
EGR_08627 |
Fatty acid-binding protein |
FP: AAACCCGTGCTTACCTTCG |
145 |
|
|
RP: CATCCGTGGTGTTCTCATCG |
|
|
EGR_09751 |
heat shock protein |
FP: TCCAGGCCGTATTACTGAGCA |
96 |
|
|
RP: ATGACTTTAGTGCCACCGT |
|
|
EGR_09229 |
LIM and SH3 domain protein |
FP: GCCAGTGTGACGTAGTCCTC |
107 |
|
|
RP: CGTTTGTGTGTTCTTCGCCAA |
|
|
EGR_00894 |
Hypothetical protein |
FP: AGTCAATTCAAGTAATCGTGCTG |
80 |
|
|
RP: TTCTCGCTTGATTTTCGCCACT |
|
|
EGR_03601 |
BoIA-like protein 3 |
FP: TTCTTAATGTCTGCCGCCAT |
104 |
|
|
RP: AGTAGAGCCTCCTTATAGTCC |
|
|
|
Control primer |
|
EGR_09068 |
Actin II |
FP: ATGGTTGGTATGGGACAAAAGG |
118 |
|
|
RP: TTCGTCACAATACCGTGCTC |
|
Table 2Sequencing data statistics
Table 2
|
Sample |
Raw reads |
Raw bases |
Clean reads |
Clean bases |
Error rate; (%) |
Q20 (%) |
Q30 (%) |
GC content (%) |
|
EgPSC_0d_1 |
60,847,206 |
9,187,928,106 |
60,032,580 |
8,950,587,025 |
0.0274 |
97.19 |
91.78 |
47.07 |
|
EgPSC_0d_2 |
65,486,882 |
9,888,519,182 |
64,718,518 |
9,648,903,196 |
0.0267 |
97.47 |
92.42 |
47.43 |
|
EgPSC_0d_3 |
44,838,914 |
6,725,837,100 |
44,709,544 |
6,679,256,433 |
0.0120 |
98.31 |
95.24 |
46.25 |
|
EgPSC_10d_1 |
50,879,460 |
7,682,798,460 |
49,912,092 |
7,433,785,290 |
0.0232 |
98.79 |
95.88 |
41.48 |
|
EgPSC_10d_2 |
45,451,476 |
6,863,172,876 |
44,642,728 |
6,648,774,193 |
0.0234 |
98.73 |
95.67 |
43.67 |
|
EgPSC_10d_3 |
51,627,418 |
7,795,740,118 |
50,541,444 |
7,440,873,654 |
0.0234 |
98.72 |
95.73 |
45.10 |
|
EgPSC_20d_1 |
62,302,076 |
9,407,613,476 |
46,103,180 |
6,618,060,319 |
0.0231 |
98.78 |
96.06 |
48.29 |
|
EgPSC_20d_2 |
56,237,888 |
8,491,921,088 |
45,097,156 |
6,377,338,828 |
0.0232 |
98.76 |
96.02 |
48.01 |
|
EgPSC_20d_3 |
54,920,334 |
8,292,970,434 |
52,009,700 |
7,464,895,752 |
0.0233 |
98.77 |
95.88 |
45.28 |
|
EgPSC_40d_1 |
59,219,662 |
8,942,168,962 |
58,640,210 |
8,770,998,130 |
0.0235 |
98.66 |
95.60 |
47.32 |
|
EgPSC_40d_2 |
52,049,106 |
7,859,415,006 |
51,576,654 |
7,738,872,659 |
0.0237 |
98.61 |
95.46 |
48.24 |
|
EgPSC_40d_3 |
54,113,540 |
8,171,144,540 |
53,665,904 |
8,042,017,580 |
0.0238 |
98.58 |
95.35 |
47.31 |
|
EgPSC_80d_1 |
49,360,298 |
7,453,404,998 |
48,784,352 |
7,308,163,088 |
0.0239 |
98.52 |
95.23 |
47.27 |
|
EgPSC_80d_2 |
49,876,008 |
7,531,277,208 |
49,296,316 |
7,387,535,278 |
0.0240 |
98.49 |
95.13 |
46.91 |
|
EgPSC_80d_3 |
50,275,646 |
7,591,622,546 |
49,770,288 |
7,457,638,729 |
0.0241 |
98.45 |
95.03 |
46.99 |
|
Average |
53,832,394 |
8,125,702,273 |
51,300,044 |
7,597,846,677 |
0.0232 |
98.46 |
95.09 |
46.44 |
Table 3COG classification of the consistently up-regulated differentially expressed genes
Table 3
|
Category |
Function |
Gene description |
|
Information storage and processing |
RNA processing and modification |
Transcription elongation regulator
Potassium voltage-gated channel subfamily C member 3 |
|
Metabolism |
Amino acid transport and metabolism |
Ornithine aminotransferase |
|
Information storage and processing |
Translation, ribosomal structure and biogenesis |
Epsin-1
NEDD8-conjugating enzyme UBE2F |
|
Poorly characterized |
Function unknown |
Synaptic vesicle membrane protein VAT-1-like protein
WD repeat-containing protein
5′-tyrosyl-DNA phosphodiesterase
Josephin-2
Cornifelin
Glycosyltransferase-like protein LARGE2
1,5-anhydro-D-fructose reductase |
|
Cellular processes and signaling |
Intracellular trafficking, secretion, and vesicular transport |
Platelet glycoprotein |
References
- 1. Zhang W, Zhang Z, Wu W, Shi B, Li J, Zhou X, Wen H, McManus DP. Epidemiology and control of echinococcosis in central Asia, with particular reference to the People’s Republic of China. Acta Trop 2015;141:235-243.
- 2. Siyadatpanah A, Anvari D, Emami Zeydi A, Hosseini SA, Daryani A, Sarvi S, Budke CM, Esmaeelzadeh Dizaji R, Mohaghegh MA, Kohansal MH, Dodangeh S, Saberi R, Gholami S. A systematic review and meta-analysis of the genetic characterization of human echinococcosis in Iran, an endemic country. Epidemiol Health 2019;41:e2019024.
- 3. Mahmoudi S, Mamishi S, Banar M, Pourakbari B, Keshavarz H. Epidemiology of echinococcosis in Iran: a systematic review and meta-analysis. BMC Infect Dis 2019;19:929.
- 4. Filippou D, Tselepis D, Filippou G, Papadopoulos V. Advances in liver echinococcosis: diagnosis and treatment. Clin Gastroenterol Hepatol 2007;5:152-159.
- 5. Xing G, Zhang H, Liu C, Guo Z, Yang X, Wang Z, Wang B, Lei Y, Yang R, Jiang Y, Lv H. Sodium arsenite augments sensitivity of Echinococcus granulosus protoscoleces to albendazole. Exp Parasitol 2019;200:55-60.
- 6. El-On J. Benzimidazole treatment of cystic echinococcosis. Acta Trop 2003;85:243-252.
- 7. Thompson RCA. Biology and systematics of Echinococcus
. Adv Parasitol 2017;95:65-109.
- 8. Albani CM, Elissondo MC, Cumino AC, Chisari A, Denegri GM. Primary cell culture of Echinococcus granulosus developed from the cystic germinal layer: biological and functional characterization. Int J Parasitol 2010;40:1269-1275.
- 9. Díaz A, Casaravilla C, Allen JE, Sim RB, Ferreira AM. Understanding the laminated layer of larval Echinococcus II: immunology. Trends Parasitol 2011;27:264-273.
- 10. Díaz A, Casaravilla C, Irigoín F, Lin G, Previato JO, Ferreira F. Understanding the laminated layer of larval Echinococcus I: Structure. Trends Parasitol 2011;27:204-213.
- 11. Ronza P, Robledo D, Bermúdez R, Losada AP, Pardo BG, Sitjà-Bobadilla A, Quiroga MI, Martínez P. RNA-seq analysis of early enteromyxosis in turbot (Scophthalmus maximus): new insights into parasite invasion and immune evasion strategies. Int J Parasitol 2016;46:507-517.
- 12. Parkinson J, Wasmuth JD, Salinas G, Bizarro CV, Sanford C, Berriman M, Ferreira HB, Zaha A, Blaxter ML, Maizels RM, Fernández C. A transcriptomic analysis of Echinococcus granulosus larval stages: implications for parasite biology and host adaptation. PLoS Negl Trop Dis 2012;6:e1897.
- 13. Liu S, Zhou X, Hao L, Piao X, Hou N, Chen Q. Genome-wide transcriptome analysis reveals extensive alternative splicing events in the protoscoleces of Echinococcus granulosus and Echinococcus multilocularis
. Front Microbiol 2017;8:1-14.
- 14. Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E, Aslett M, Beasley H, Bennett HM, Cai X, Camicia F, Clark R, Chcher M, De Silva N, Day TA, Deplazes P, Estrada K, Fernández C, Holland PWH, Hou J, Hu S, Huckvale T, Hung SS, Kamenetzky L, Keane JA, Kiss F, Koziol U, Lambert O, Liu K, Luo X, Luo Y, Macchiaroli N, Nichol S, Paps J, Parkinson J, Pouchkina-Stantcheva N, Riddiford N, Rosenzvit M, Salinas G, Wasmuth JD, Zamanian M, Zheng Y, Cai J, Soberón X, Olson PD, Laclette JP, Brehm K, Berriman M. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 2013;496:57-63.
- 15. Debarba JA, Monteiro KM, Moura H, Barr JR, Ferreira HB, Zaha A. Identification of newly synthesized proteins by Echinococcus granulosus protoscoleces upon induction of strobilation. PLoS Negl Trop Dis 2015;9:1-18.
- 16. Liu C, Yin J, Xue J, Tao Y, Hu W, Zhang H.
In vitro effects of amino alcohols on Echinococcus granulosus
. Acta Trop 2018;182:285-290.
- 17. Elissondo MC, Dopchiz MC, Brasesco M, Denegri G.
Echinococcus granulosus: first report of microcysts formation from protoscoleces of cattle origin using the in vitro vesicular culture technique. Parasite 2004;11:415-418.
- 18. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009;25:1754-1760.
- 19. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions, and gene fusions. Genome Biol 2013;14:R36.
- 20. Zhu Y, Cao X, Zhang X, Chen Q, Wen L, Wang P. DNA methylation-mediated klotho silencing is an independent prognostic biomarker of head and neck squamous carcinoma. Cancer Manag Res 2019;11:1383-1390.
- 21. Lateef A, Prabhudas SK, Natarajan P. RNA sequencing and de novo assembly of Solanum trilobatum leaf transcriptome to identify putative transcripts for major metabolic pathways. Sci Rep 2018;8:15375.
- 22. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26:136-138.
- 23. He JJ, Ma J, Elsheikha HM, Song HQ, Huang SY, Zhu XQ. Transcriptomic analysis of mouse liver reveals a potential hepato-enteric pathogenic mechanism in acute Toxoplasma gondii infection. Parasit Vectors 2016;9:1-13.
- 24. Phuphisut O, Ajawatanawong P, Limpanont Y, Reamtong O, Nuamtanong S, Ampawong S, Chaimon S, Dekumyoy P, Watthanakulpanich D, Swierczewski BE, Adisakwattana P. Transcriptomic analysis of male and female Schistosoma mekongi adult worms. Parasit Vectors 2018;11:504.
- 25. Fen W, Xunuo L, Hongye W, Kai L, Junjie F, Bin Y. Identification of optimal reference genes during development of hydatid cysts from protoscoleces and transcriptional expression of candidate aquaporins. J Third Mil Med Univ 2018;40:1533-1541. (in Chinese).
- 26. Yoo WG, Kim TI, Li S, Kwon OS, Cho PY, Kim TS, Kim K, Hong SJ. Reference genes for quantitative analysis on Clonorchis sinensis gene expression by real-time PCR. Parasitol Res 2009;104:321-328.
- 27. Guo B, Zhang Z, Zheng X, Guo Y, Guo G, Zhao L, Ren C, Wang B, Yang M, Shou X, Zhang W, Jia B. Prevalence and molecular characterization of Echinococcus granulosus sensu stricto in Northern Xinjiang, China. Korean J Parasitol 2019;57:153-159.
- 28. Mandal S, Deb Mandal M. Human cystic echinococcosis: epidemiologic, zoonotic, clinical, diagnostic and therapeutic aspects. Asian Pac J Trop Med 2012;5:253-60.
- 29. Daneshpour S, Kefayat A, Mofid M, Rostami Rad S, Yousofi Darani H. Effect of hydatid cyst fluid antigens on induction of apoptosis on breast cancer cells. Adv Biomed Res 2019;8:27.
- 30. Mandal S, Deb Mandal M. Human cystic echinococcosis: epidemiologic, zoonotic, clinical, diagnostic and therapeutic aspects. Asian Pac J Trop Med 2012;5:253-260.
- 31. Dezaki ES, Yaghoubi MM, Spiliotis M. Comparison of ex vivo harvested and in vitro cultured materials from Echinococcus granulosus by measuring expression levels of five genes putatively involved in the development and maturation of adult worms. Parasitol Res 2016;115:4405-4416.
- 32. Huang Y, Xiong JL, Gao XC, Sun XH. Transcriptome analysis of the Chinese giant salamander (Andrias davidianus) using RNA-sequencing. Genomics Data 2017;14:126-131.
- 33. Zheng H, Zhang W, Zhang L, Zhang Z, Li J, Lu G, Zhu Y, Wang Y, Huang Y, Liu J, Kang H, Chen J, Wang L, Chen A, Yu S, Gao Z, Jin L, Gu W, Wang Z, Zhao L, Shi B, Wen H, Lin R, Jones MK, Brejova B, Vinar T, Zhao G, McManus DP, Chen Z, Zhou Y, Wang S. The genome of the hydatid tapeworm Echinococcus granulosus
. Nat Genet 2013;45:1168-1175.
- 34. Almeida GT, Amaral MS, Beckedorff FCF, Kitajima JP, DeMarco R, Verjovski-Almeida S. Exploring the Schistosoma mansoni adult male transcriptome using RNA-seq. Exp Parasitol 2012;132:22-31.
- 35. Li WH, Zhang NZ, Yue L, Yang Y, Li L, Yan HB, Li TT, Qu ZG, Jia WZ, Fu BQ. Transcriptomic analysis of the larva Taenia multiceps
. Res Vet Sci 2017;115:407-411.
- 36. Camicia F, Celentano AM, Johns ME, Chan JD, Maldonado L, Vaca H, Di Siervi N, Kamentezky L, Gamo AM, Ortega-Gutierrez S, Martin-Fontecha M, Davio C, Marchant JS, Rosenzvit MC. Unique pharmacological properties of serotoninergic G-protein coupled receptors from cestodes. PLoS Negl Trop Dis 2018;12:1-27.
- 37. de Saram PSR, Ressurreição M, Davies AJ, Rollinson D, Emery AM, Walker AJ. Functional mapping of protein kinase A reveals its importance in adult Schistosoma mansoni motor activity. PLoS Negl Trop Dis 2013;7:e1988.
- 38. Vorontsova YL, Slepneva IA, Yurlova NI, Ponomareva NM, Glupov VV. The effect of trematode infection on the markers of oxidative stress in the offspring of the freshwater snail Lymnaea stagnalis
. Parasitol Res 2019;118:3561-3564.
- 39. Ty MC, Zuniga M, Götz A, Kayal S, Sahu PK, Mohanty A, Mohanty S, Wassmer SC, Rodriguez A. Malaria inflammation by xanthine oxidase-produced reactive oxygen species. EMBO Mol Med 2019;11:e9903.
- 40. Sayed AA, Williams DL. Biochemical characterization of 2-cys peroxiredoxins from Schistosoma mansoni
. J Biol Chem 2004;279:26159-26166.
- 41. Cancela M, Paes JA, Moura H, Barr JR, Zaha A, Ferreira HB. Unraveling oxidative stress response in the cestode parasite Echinococcus granulosus
. Sci Rep 2019;9:15876.
- 42. Zalewska A, Ziembicka D, Żendzian-Piotrowska M, Maciejczyk M. The Impact of high-fat diet on mitochondrial function, free radical production, and nitrosative stress in the salivary glands of wistar rats. Oxid Med Cell Longev 2019;2019:1-15.
- 43. Ozcan C, LI Z, Kim G, Jeevanandam V, Uriel N. Molecular mechanism of the association between atrial fibrillation and heart failure includes energy metabolic dysregulation due to mitochondrial dysfunction. J Card Fail 2019;25:911-920.
- 44. Li Y, Li P, Peng Y, Wu Q, Huang F, Liu X, Li X, Zhou H, Guo D, Shi D, Zhou XN, Fan X. Expression, characterization and crystal structure of thioredoxin from Schistosoma japonicum
. Parasitology 2015;142:1044-1052.
- 45. Wilkinson SR, Horn D, Prathalingam SR, Kelly JM. RNA interference identifies two hydroperoxide metabolizing enzymes that are essential to the bloodstream form of the African trypanosome
. J Biol Chem 2003;278:31640-31646.
- 46. Bombaça ACS, Brunoro GVF, Dias-Lopes G, Ennes-Vidal V, Carvalho PC, Perales J, d’Avila-Levy CM, Valente RH, Menna-Barreto RFS. Glycolytic profile shift and antioxidant triggering in symbiont-free and H2O2-resistant Strigomonas culicis
. Free Radic Biol Med 2020;146:392-401.
- 47. Lopez-Gonzalez V, La-Rocca S, Arbildi P, Fernandez V. Characterization of catalytic and non-catalytic activities of EgGST2-3, a heterodimeric glutathione transferase from Echinococcus granulosus
. Acta Trop 2018;180:69-75.
- 48. Gaba S, Jamal S; Open Source Drug Discovery Consortium, Scaria V. Cheminformatics models for inhibitors of Schistosoma mansoni thioredoxin glutathione reductase. Sci World J 2014;2014:1-9.
- 49. do Santos GB, Monteiro KM, da Silva ED, Battistella ME, Ferreira HB, Zaha A. Excretory/secretory products in the Echinococcus granulosus metacestode: is the intermediate host complacent with infection caused by the larval form of the parasite? Int J Parasitol 2016;46:843-856.
- 50. Inokuma K, Kurono H, den Haan R, van Zyl WH, Hasunuma T, Kondo A. Novel strategy for anchorage position control of GPI-attached proteins in the yeast cell wall using different GPI-anchoring domains. Metab Eng 2020;57:110-117.
- 51. Phung LT, Chaiyadet S, Hongsrichan N, Sotillo J, Dieu HDT, Tran CQ, Brindley PJ, Loukas A, Laha T. Recombinant Opisthorchis viverrini tetraspanin expressed in Pichia pastoris as a potential vaccine candidate for opisthorchiasis. Parasitol Res 2019;118:3419-3427.
- 52. Tomii K, Santos HJ, Nozaki T. Genome-wide analysis of known and potential tetraspanins in Entamoeba histolytica
. Genes (Basel) 2019;10:E885.
- 53. Coakley G, Maizels RM, Buck AH. Exosomes and other extracellular vesicles: the new communicators in parasite infections. Trends Parasitol 2015;31:477-489.
- 54. Siles-Lucas M, Sánchez-Ovejero C, González-Sánchez M, González E, Falcón-Pérez JM, Boufana B, Fratini F, Casulli A, Manzano-Román R. Isolation and characterization of exosomes derived from fertile sheep hydatid cysts. Vet Parasitol 2017;236:22-33.
- 55. Park EJ, Kwon TH. A Minireview on vasopressin-regulated aquaporin-2 in kidney collecting duct cells. Electrolytes Blood Press 2015;13:1.
- 56. Horgan CP, Hanscom SR, Jolly RS, Futter CE, McCaffrey MW. Rab11-FIP3 binds dynein light intermediate chain 2 and its overexpression fragments the Golgi complex
. Biochem Biophys Res Commun 2010;394:387-392.
- 57. Fox AR, Maistriaux LC, Chaumont F. Toward understanding of the high number of plant aquaporin isoforms and multiple regulation mechanisms. Plant Sci 2017;264:179-187.
- 58. Huang Y, Li W, Lu W, Xiong C, Yang Y, Yan H, Liu KC, Cao P. Cloning and in vitro characterization of a Schistosoma japonicum aquaglyceroporin that functions in osmoregulation. Sci Rep 2016;6:1-8.
- 59. Faghiri Z, Camargo SM, Huggel K, Forster IC, Ndegwa D, Verrey F, Skelly PJ. The tegument of the human parasitic worm Schistosoma mansoni as an excretory organ: the surface aquaporin SmAQP is a lactate transporter. PLoS One 2010;5:e10451.