Abstract
The Japanese amberjack Seriolae quinqueradiata is one of the most consumed fish species among the Koreans. However, information regarding parasitic infection in Japanese amberjack is scarce. This study described the morphological and molecular characteristics of a species of philometrid nematode, Philometroides seriolae, which was recovered from Japanese amberjack. This fish was caught in the sea of Goseong-gun, Gangwon-do, Republic of Korea (Korea). Six P. seriolae (Nematoda: Philometridae) were recovered from 2 Japanese amberjacks. These parasites were subgravid female which were 325–420 mm long and 2.95–3.27 mm wide. Furthermore, they had typical papillae distributed on their body surface with 14 papillae at the apical view. Sequence analysis of the small subunits of ribosomal RNA (SSU rRNA) showed high sequence identity (99.8%, 1,607/1,611-bp) with that of P. seriolae (GenBank accession no. FJ155811). This nematode species has been newly added to the Korean nematode fauna.
-
Key words: Seriola quinqueradiata, Philometroides seriolae, small subunit rRNA
The Japanese amberjack
Seriola quinqueradiata (Temminck & Shlegel, 1845) is a marine fish species belonging to the family Carangidae. This fish is distributed in the neritic seas of the northwestern Pacific extending along the South and East Sea of the Republic of Korea (Korea), Japan (except for the Southern Ryukyu Islands), and circling to Hawaii. It can grow up to a length of 150 cm and weigh 40 kg [
1]. This fish is of economic importance owing to its large volume of flesh and good taste. It is cultured both in Korea and Japan [
2]. In Korea, this species is widely consumed by people as various recipes. Most commonly is eaten raw during the winter season (December to February).
Philometroides seriolae (Ishii, 1931) Yamaguti, 1935 is a tissue parasitic nematode of Japanese amberjack. In Japan, this parasite is associated with severe economic losses in cultured Japanese amberjack [
2,
3]. This parasite is also popular within the Korean fishing industry, sushi restaurants, and people who enjoy sushi. Despite its reputation, this species of parasite has not been recorded and identified in a scientific manner in Korea. Therefore, the purpose of the present study was to provide scientific evidence of
P. seirolae in Korea based on its morphological and molecular characterization.
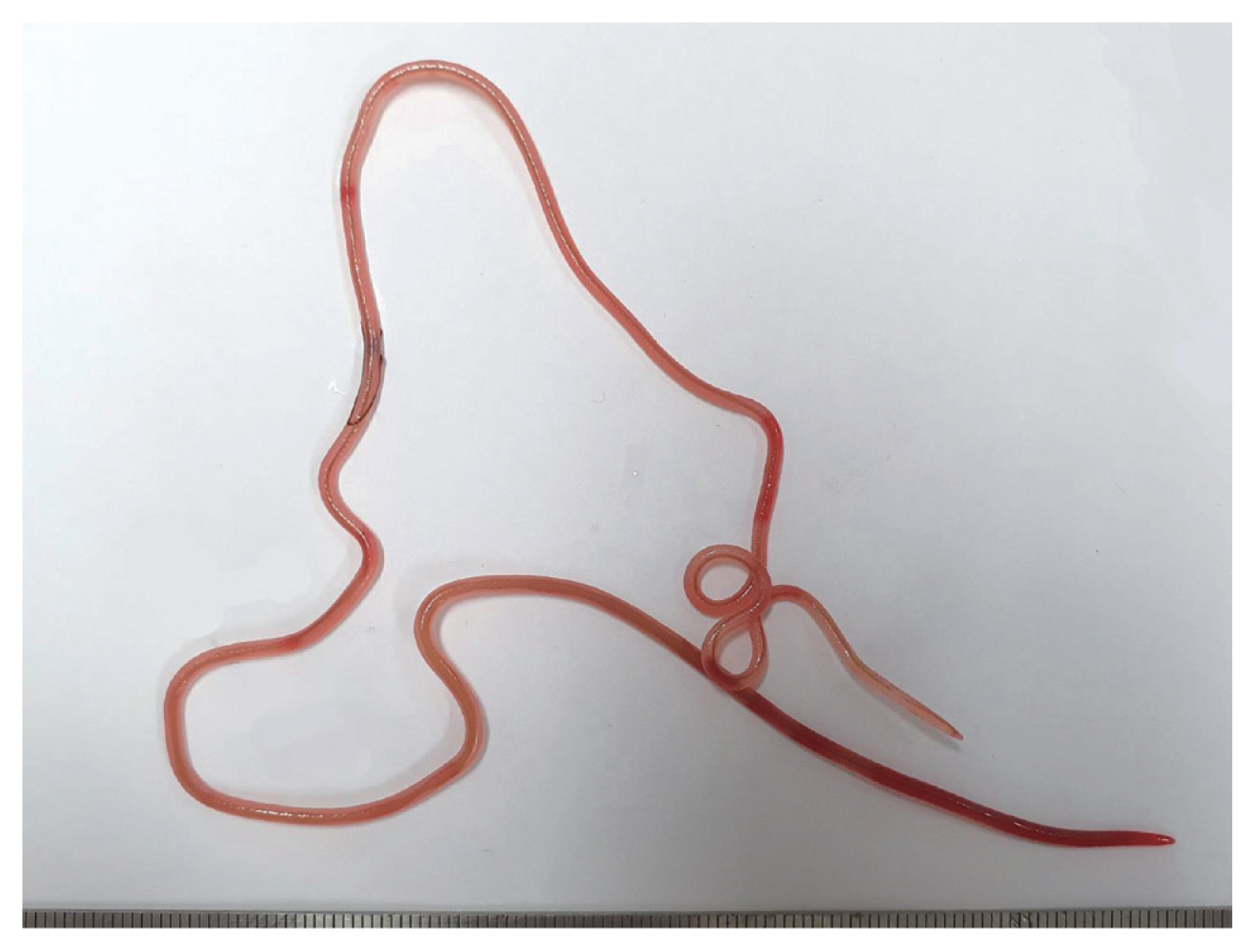
In December 2019, 2 Japanese amberjacks were purchased and examined for helminth parasites. The fishes were caught off the coast of Goseong-gun (county) at Gangwon-do (province) in Korea. They weighed 9.6 kg and 10.4 kg, respectively. However, their size and gender were not recorded. The fish were prepared by following the common procedure for sashimi. The parasite specimens were recovered from the muscle areas using naked eyes (
Fig. 1). A total of 6 specimens (4 and 2 worms, respectively) were collected. These were observed under a light microscope after clearing with a lacto-phenol solution. All measurements were done in micrometers, unless otherwise indicated. The voucher specimens were deposited in the National Institute of Biological Resources (NIBR) located in Incheon, Korea (specimen codes NIBRIV0000880102 and NIBRIV0000880103), and the Department of Parasitology, School of Medicine, Chungbuk National University, Cheongju, Korea.
Following the protocol of manufacturer, the genomic DNA was extracted using Chelex (Sigma-Aldrich, St. Louis, Missouri, USA) from the parasite tissues of adult worms without fixation. Polymerase chain reaction (PCR) was conducted to amplify the target gene, small subunit ribosomal RNA (SSU rRNA), using a GeneAmp PCR system 9700 (Applied Biosystems, Langen, Germany). The PCR conditions and primer sets followed the protocols of a previous study on philometrids [
4]. The PCR products were purified using a gel extraction kit (Qiagen Ltd., Valencia, California, USA). The products were then sequenced using the Applied Biosystems 3730xl DNA Analyzer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) equipped in Cosmogenetech, Daejeon, Korea, with the same primer sets as those used for PCR [
4]. For molecular comparisons, the obtained sequences were assembled and edited using ClustalW [
5], Mega7 [
6], and BLASTn analysis (
http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic trees of maximum likelihood (ML) and Bayesian inference (BI) were constructed based on the selected SSU rRNA sequences of marine philometrids provided from the GenBank database (
Table 1). The analyses were performed using the GTR+I+G model implemented in jModeltest 2.1.10 [
7]). The ML and BI trees were calculated using PhyML [
8] and MrBayes 3.2.6 [
9]. The bootstrap and
P-values were discarded when they represented lower than 70% in ML and 0.95 in BI.
Family Philometridae Baylis and Daubney, 1926
Genus Philometroides Yamaguti, 1935
Species
Philometroides seriolae (Ishii, 1931) Yamaguti, 1935 (
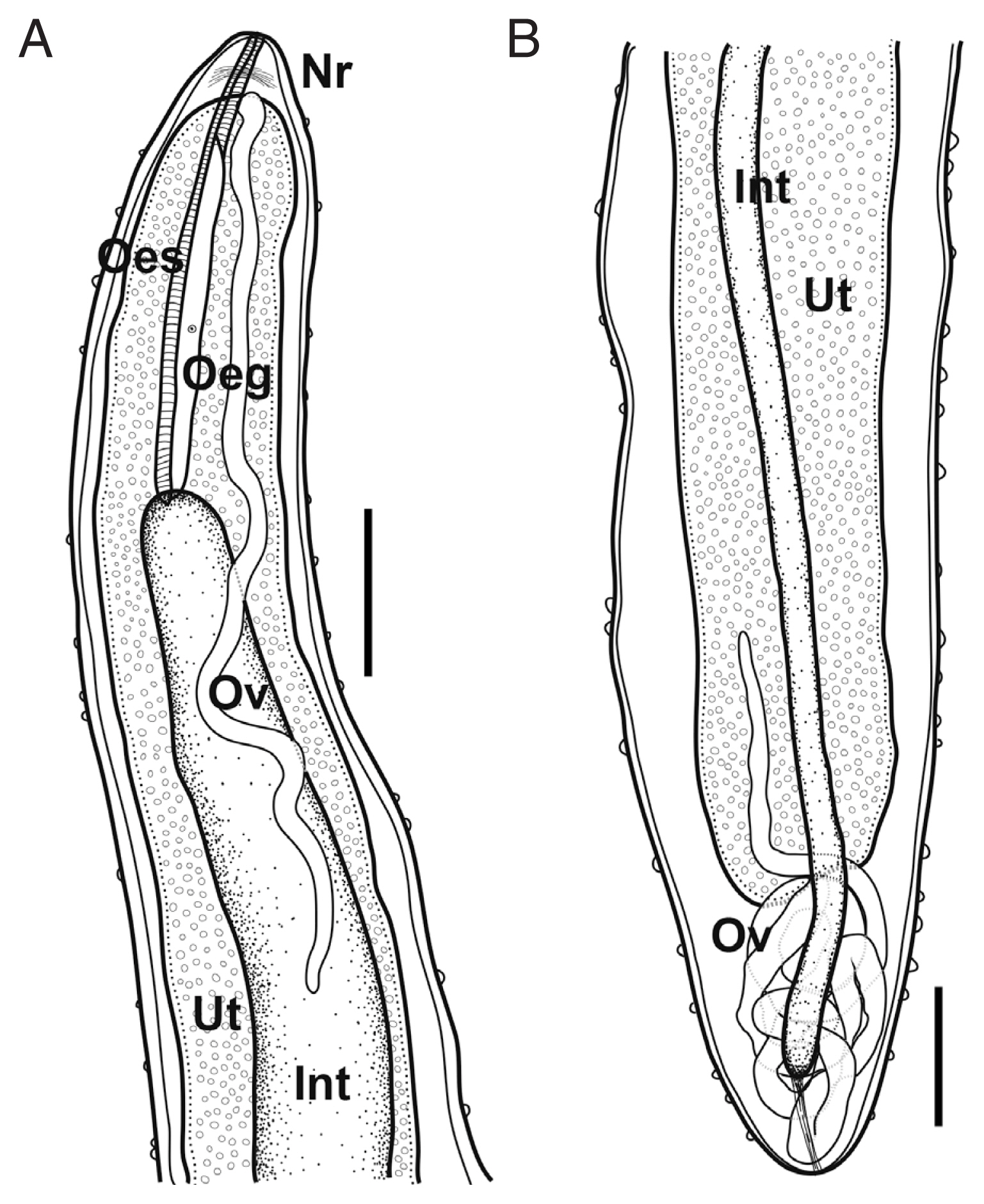
Fig. 2).
Based on 3 subgravid females. Body pinkish to reddish, subcylindrical, uniformly broad. Anterior and posterior extremity blunt. Anterior end somewhat narrowed. Length of body 325–420 mm, maximum width 2.95–3.27 mm; maximum body width/body length 1: 110–135. Cuticle thick, numerous papilliform protuberances, individually dotted on body surface, 8–15 μm high in anterior part, 9–32 μm in posterior. Cephalic papillae very small, not well detected in some specimens, located near oral opening, arranged in inner and outer circles; 4 smaller submedian and 2 lateral papillae (inner circle); 4 larger submedian pairs of papillae with pairs of amphids.
Oral opening triangular, connected into slightly swollen anterior bulb of oesophagus. Anterior swelling of oesophagus 191–209 μm long, 155–191 μm wide. Following portion of oesophagus cylindrical, musculated, 4.73–5.48 mm long, 0.43–0.61 mm in maximum width. Occupying 1.1–1.7% of body length. Dorsal oesophageal gland well developed, issued from the level of nerve ring to posterior extremity of oesophagus. Cell nucleus of oesophageal gland weakly developed, located at mid-level of gland. Nerve ring located 201–397 μm from anterior extremity. Small oesophageal ventriculus projected into intestine. Intestine long, blind, somewhat light green when observed under light microscope, connected to posterior end of body. Posterior part of intestine somewhat gradually narrowed posteriorly, connected to posterior extremity of body with strong fibrous cord-like ligament, ended 400–600 μm from posterior extremity.
Ovaries long, narrow, convoluted, issued from each extremity of body, connected to uterus. Uterus expanded, occupying most of body space except near regions of each anterior and posterior extremity, filled with embryos and embryonated eggs.
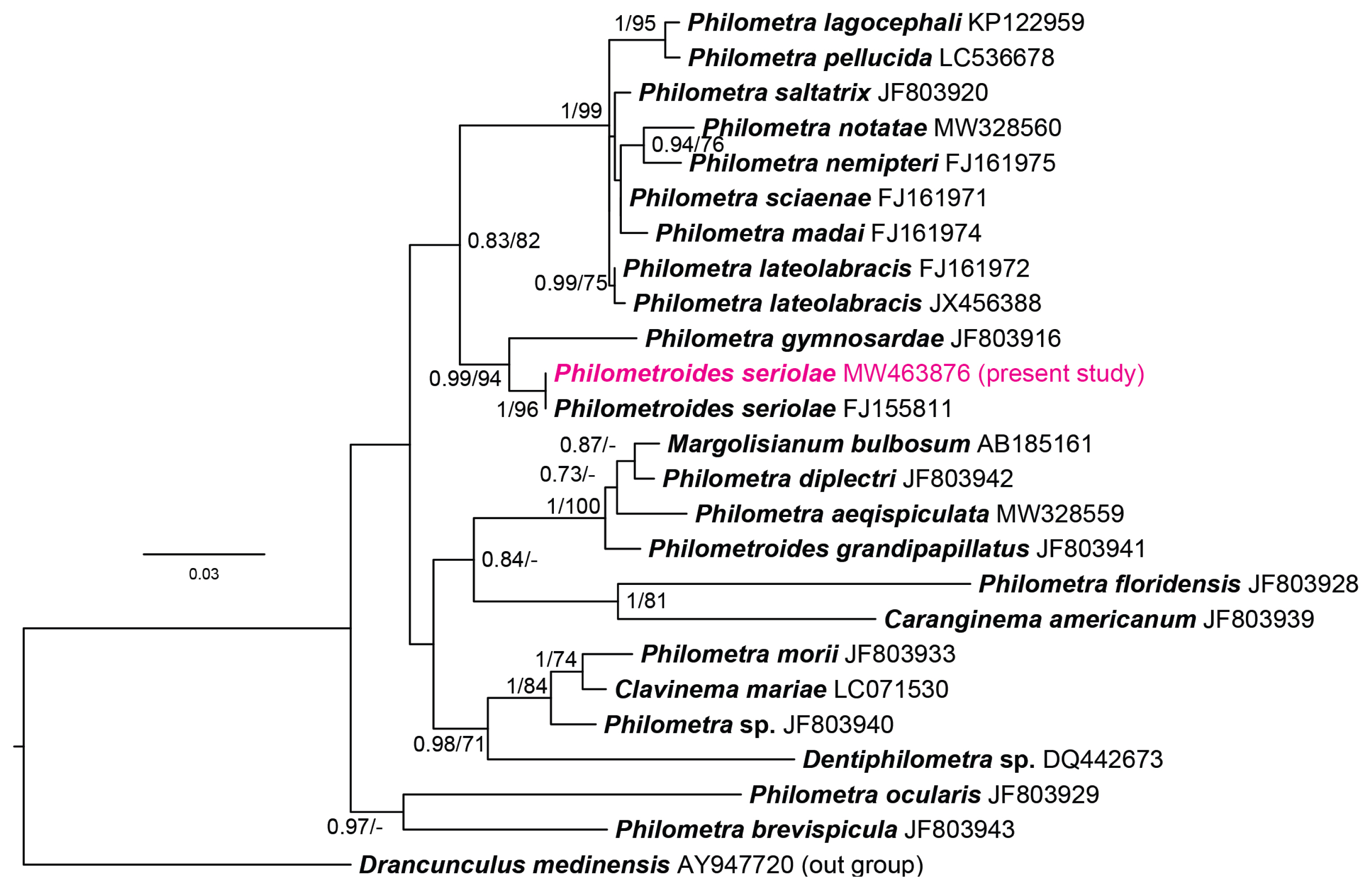
The molecular analysis revealed that both parasitic specimens obtained from fish hosts showed 100% identical sequences of partial SSU rRNA (1,624/1,624 bp). BLASTn analysis indicated that sequences obtained from philometrid nematodes from the Japanese amberjacks revealed high sequence identity (99.8%, 1,607/1,611) with a sequence of P. seriolae provided in GenBank (FJ155811). The sequence newly generated from the present study was submitted to the GenBank database under the accession number MW463876. In the phylogenetic analyses, both ML and BI trees represented the newly generated sequence and formed a clade with the provided P. seriolae sequence generated from P. seriolae in Japan (FJ155811) with a strong support from node values (1/96).
These results indicate that the specimens of this study can be considered to be
P. seriolae. Yamaguti suggested the genus
Philomtroides based on the presence of cuticular bosses all over the body surface with strongly developed parietal muscles [
10]. Due to the distinct morphological differences with the genus
Philometra Costa, 1845, philometrid nematodes equipped with cuticular bosses have been classified as genus
Philometroides. However, the current results based on phylogenetic analysis revealed that cuticular bosses might not be a major key to identify the genus [
4]. However, here the genus was retained until the taxonomic status could be revised.
Based on morphological observations and molecular comparison, the specimens collected from Japanese amberjacks were identified as
P. seriolae. The present specimens were equipped with typical papillae distributed on their body surface, which are key characteristics of the genus
Philometroides [
10]. The morphological features (the arrangement of organs and their measurements) seemed well fitted with the morphological characteristics of
P. seriolae as described by previous authors [
2–
4,
10]. Moravec and Ogawa [
2] mentioned that subgravid females show 14 papillae with a pair of amphids arranged in 2 circles. Unlike their observations, Quiazon et al. [
4] observed gravid females and mentioned that they were equipped with only 8 small cephalic papillae without amphids arranged in a circle. The present study observed 14 small papillae with pairs of amphids in 2 circles, similar to the observations by Moravec and Ogawa [
2]. They suspected that the morphological differences described by authors were due to differences in developmental stages (subgravid vs. gravid) and reasoned them to be atrophied in large gravid females. All the specimens in the present study were subgravid females with their uterus filled with larvae and eggs together. This should be confirmed in further studies by observations of fully gravid females.
This long reddish worm is found in the flesh of Japanese amberjack. It has been generally suspected as P. seriolae among sushi-enjoyers, chefs, and fishermen in Korea. It has been frequently mentioned by both experts and general public using social network media and online video platforms. However, it was not confirmed by scientific identification, but it was identified according to the literature and by comparing its gross shape (personal communication). Thus, this study is the first in Korea to identify P. seriolae using morphological and molecular analysis. It is presumed that the Japanese amberjacks living in Korean offshore waters are infected with a high infection rate and population, which needs to be confirmed in a subsequent study.
According to the testimonies, the reason why Koreans avoid to consuming Japanese amberjack in the summer season is that there are many tissue-parasitic worms and lower fat contents that induce Japanese amberjack to lose flesh and tastes together. When fishes are purchased in Korea, they are generally priced based on their weight. Thus, if the number of parasites in flesh and the affected areas increase largely, it leads to economic losses, as in cases of fish cultures in Japan [
2]. The seasonal differences of parasitic infections in Japanese amberjack are thought to be closely related to their life history. Nakajima et al. [
11,
12] recorded marine copepods as the first intermediate hosts and observed the reproductive emergence of
P. seriolae from the fish host. The season of emergence was from the middle of August to the beginning of September [
12]. Although the present study described the first finding of
P. seriolae from Korean sea, the study period was limited to winter season. For a better understanding of their life cycle and the impact on the fisheries industry, the further study should be carried out in consideration of the seasonal differences. And it may allowed to get additional information such as the male worms who are not well known [
2].
In conclusion, this study proved that the philometrid parasite, P. seriolae, is distributed in the seas of Korea, and further studies may elucidate their prevalence in fishes, flesh yield of fishes after removing parasites, and their potential risks to humans.
Notes
-
The authors declare no conflict of interest related to this study.
ACKNOWLEDGMENT
This work was supported by a grant from the National Institute of Biological Resources (NIBR), which is funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR no. 2020-02-204).
Fig. 1A gross female specimen of Philometroides seriolae (Ishii, 1931) recovered in a Japanese amberjack, Seriolae quinqueradiata (Temminck & Shlegel, 1845), from Goseong-gun, Gangwon-do, Korea.
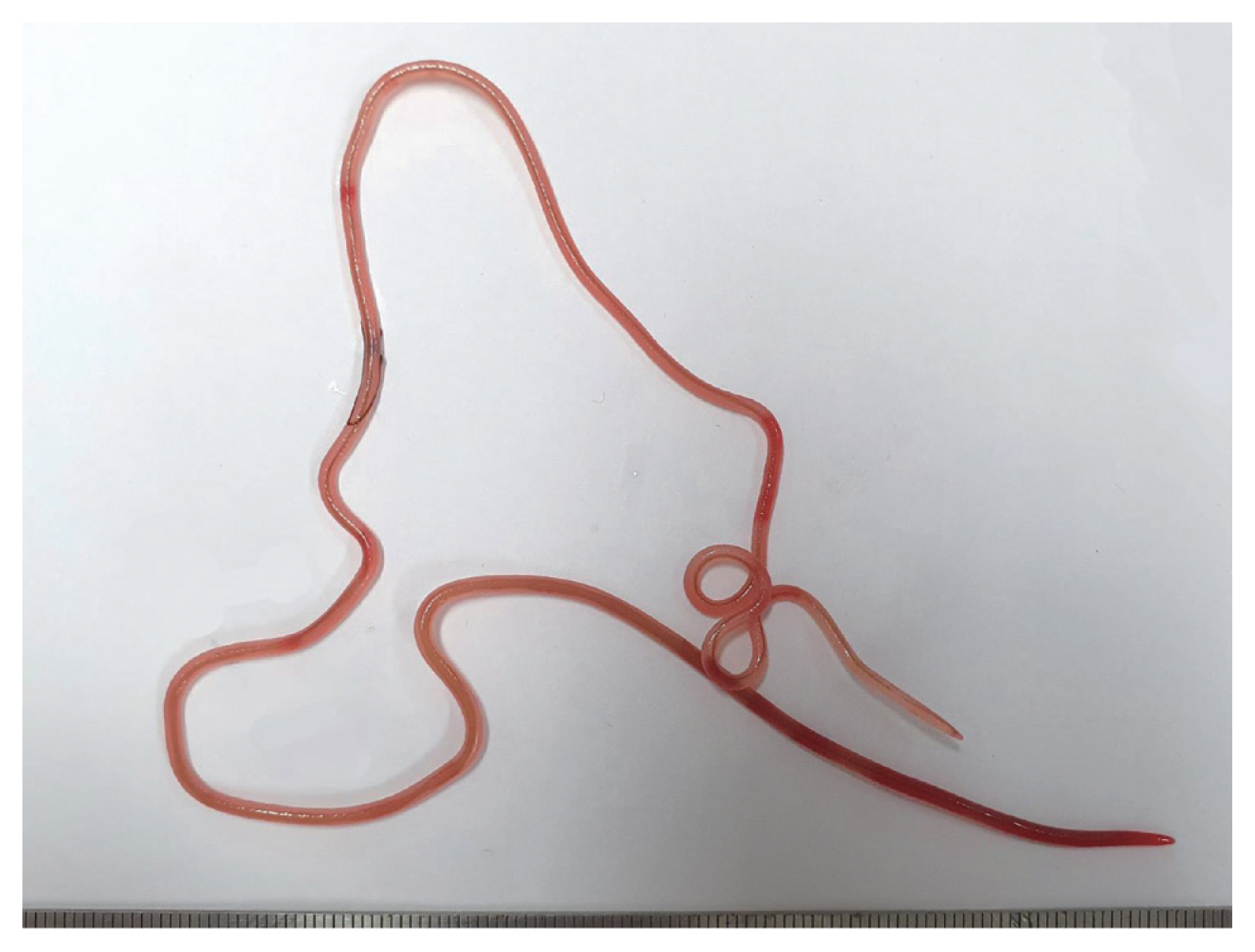
Fig. 2Schematic drawings of Philometroides seriolae (Ishii, 1931) subgravid female. (A) Anterior extremity. (B) Posterior extremity. Bar= 2 mm. Oeg, esophageal gland; Int, intestine; Nr, nerve ring; Oes, oesophagus; Ov, ovary; Ut, uterus.
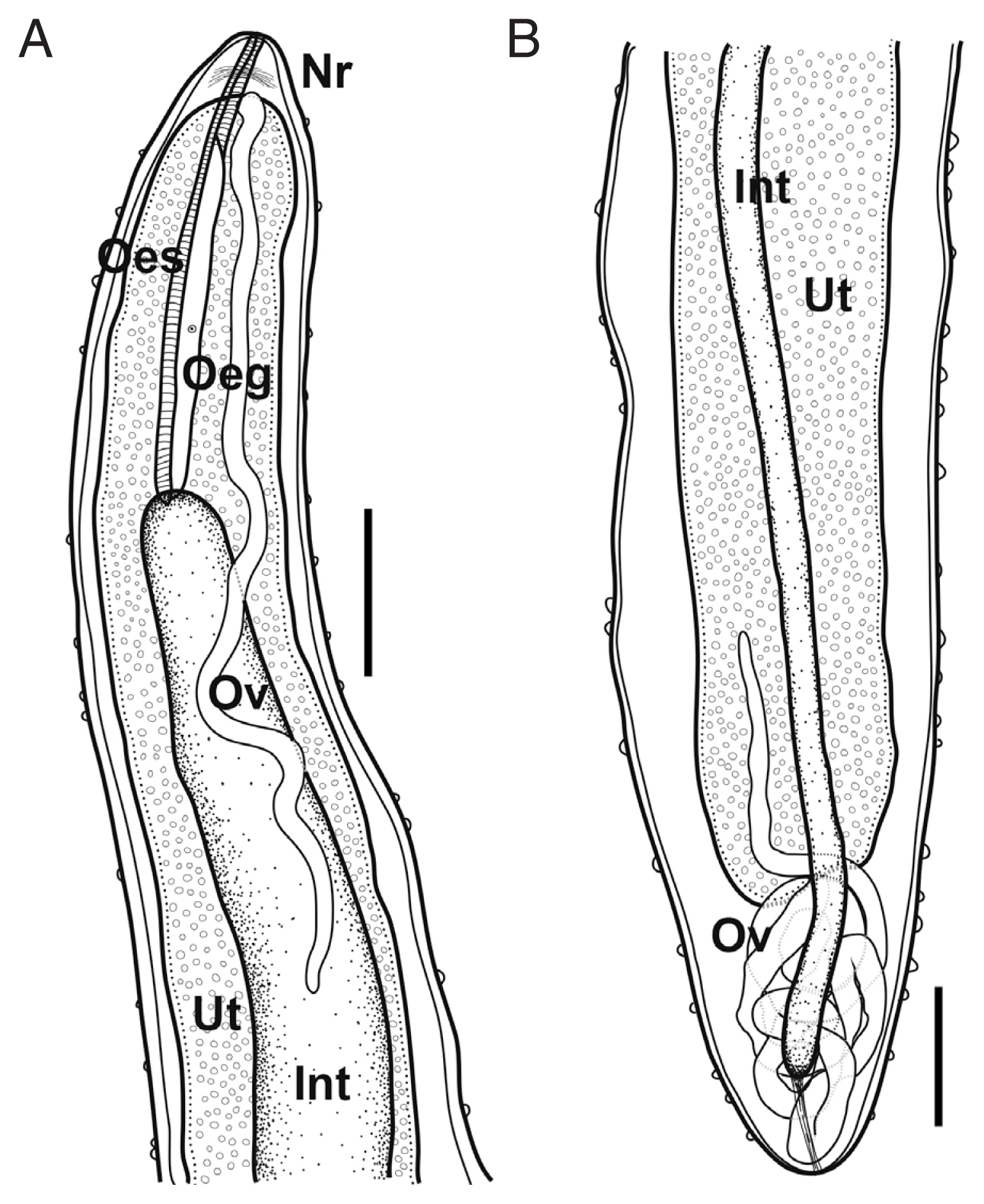
Fig. 3Bayesian tree constructed based on the phylogenetic analyses of the selected SSU rRNA sequences dataset of marine philometrid nematodes. The node values represent the Bayesian posterior probability (100,000 generations with discarding primal 25%) and maximum likelihood bootstrap value (1,000 replications).

Table 1Selected sequences used for phylogenetic analyses in the present study. Information of parasite and host species with locality were noted
Table 1
|
Parasite species |
Host species |
Locality |
GenBank accession no. |
|
Philometridae |
|
Caranginema americanum
|
Caranx hippos
|
USA |
JF803939 |
|
Clavinema mariae
|
Sebastes schlegelii
|
Korea |
LC071530 |
|
Dentiphilometra sp. |
Lutjanus griseu
|
Mexico |
DQ442673 |
|
Margolisianum bulbosum
|
Paralichthys lethostigma
|
USA |
AB185161 |
|
Philometra aequispiculata
|
Strongylura notata
|
USA |
MW328559 |
|
Philometra brevispicula
|
Lutjanus griseu
|
USA |
JF803943 |
|
Philometra diplectri
|
Diplectrum formosum
|
USA |
JF803942 |
|
Philometra floridensis
|
Sciaenops ocellatus
|
USA |
JF803928 |
|
Philometra gymnosardae
|
Gymnosarda unicolor
|
Maldives |
JF803916 |
|
Philometra lagocephali
|
Lagocephalus lunaris
|
China |
KP122959 |
|
Philometra lateolabracis
|
Lateolabrax japonicus
|
Japan |
FJ161972 |
|
Philometra lateolabracis
|
Epinephelus costae
|
Turkey |
JX456388 |
|
Philometra madai
|
Pagrus major
|
Japan |
FJ161974 |
|
Philometra morii
|
Epinephelus morio
|
USA |
JF803933 |
|
Philometra nemipteri
|
Nemipterus virgatus
|
Japan |
FJ161975 |
|
Philometra notatae
|
Strongylura notata
|
USA |
MW328560 |
|
Philometra ocularis
|
Epinephelus areolatus
|
New Caledonia |
JF803929 |
|
Philometra pellucida
|
Arothron nigro
|
Australia |
LC536678 |
|
Philometra saltatrix
|
Pomatomus saltatrix
|
USA |
JF803920 |
|
Philometra sciaenae
|
Pennahia argentata
|
Japan |
FJ161971 |
|
Philometra sp. |
Mycteroperca microlepis
|
USA |
JF803940 |
|
Philometroides grandipapillatus
|
Caranx hippos
|
USA |
JF803941 |
|
Philometroides seriolae
|
Seriola quinqueradiata
|
Japan |
FJ155811 |
|
Philometroides seriolae
|
Seriola quinqueradiata
|
Korea |
MW463876*
|
|
|
Dracunculidae (out group) |
|
Dracunculus medinensis
|
Homo sapiens
|
Ghana |
AY947720 |
References
- 1. Froese R, Pauly D. FishBase; World Wide Web electronic publication; 2019. http://www.fishbase.org
- 2. Moravec F, Ogawa K. Description of two new species of Philometra Costa, 1845 (Nematoda: Philometridae) from marine fishes off Japan, with notes on Philometroides seriolae (Yamaguti, 1935). Acta Parasitol 2019;64:829-838. http://doi.org/10.2478/s11686-019-00107-y
- 3. Moravec F, Nagasawa K, Ogawa K. Observations on five species of philometrid nematodes from marine fishes in Japan. Syst Parasitol 1998;40:67-80. http://doi.org/10.1023/A:1005976012723
- 4. Quiazon KMA, Yoshinaga T, Doi H, Araki J, Ogawa K. First description of male Philometra thaiensis Moravec, Fiala et Dyková, 2004 (Nematoda: Philometridae) from the body cavity of the eyespot pufferfish Tetradon biocellatus Tirant, and evolutionary relationships of this species with other dracunculoids as inferred from SSU rRNA gene sequences. Helminthol 2014;51:236-245. http://doi.org/10.2478/s11687-014-0235-6
- 5. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 2003;31:3497-3500. http://doi.org/10.1093/nar/gkg500
- 6. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725-2729. http://doi.org/10.1093/molbev/mst197
- 7. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 2012;9:772. https://doi.org/10.1038/nmeth.2109
- 8. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010;59:307-321. https://doi.org/10.1093/sysbio/syq010
- 9. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61:539-542. https://doi.org/10.1093/sysbio/sys029
- 10. Yamaguti S. Studies on the helminth fauna of Japan. Part 9. Nematodes of fishes, 1. Jpn J Zool 1935;6:337-386.
- 11. Nakajima K, Egusa S. Studies on the life history of Philometroides seriolae (Ishii, 1931) Yamaguti, 1935 (Nematode)–I: experimental infection of copepods with the first stage larva. Fish Pathol 1970;5:1-3. (in Japanese). https://doi.org/10.3147/jsfp.5.12
- 12. Nakajima K, Egusa S, Nakajima H. Reproductive emergence of Philometroides seriolae from the host. Fish Pathol 1970;4:83-86. (in Japanese). https://doi.org/10.3147/jsfp.4.83