Abstract
Laelapinae mites are involved in transmission of microbial diseases between wildlife and humans, with an impact on public health. In this study, 5 mite members in the subfamily Laelapinae (laelapin mites; LM) were morphologically identified by light microscopy, and the phylogenetic relationship of LM was analyzed in combination with the sequence information of part of the LM cytochrome oxidase subunit I (cox1) gene. The morphological identification revealed that 5 mites belonged to the genera Laelaps and Haemolaelaps, respectively. Sequence analysis showed that the ratio of non-synonymous mutation rate to synonymous mutation rate of LM was less than 1, indicating that the LM cox1 gene had undergone purifying selection. Phylogenetic analysis showed that the Laelapinae is a monophyletic group. The genera Haemolaelaps and Hyperlaelaps did not separated into distinct clades but clustered together with species of the genus Laelaps. Our morphological and molecular analyses to describe the phylogenetic relationships among different genera and species of Laelapinae provide a reference for the improvement and revision of the LM taxonomy system.
-
Key words: Laelapinae, morphological identification, phylogenetic analysis, cox1
INTRODUCTION
Laelapinae belongs to the family Laelapidae, 65 species in 35 genera have been reported in China [
1]. Laelapinae mites are common ectoparasites of small mammals, especially rodents [
2]. The basic process of the development of Laelapinae mites (LM) can be divided into 5 stages: egg, larva, first nymph, second nymph, and adult. They complete the majority of their life cycle in the nest of the host [
3]. Adult female mites occur mostly on the host, while adult males (and other life stages) remain primarily in the nests of hosts [
4]. Some LM are obligate, non-exclusive haematophages [
5] (they feed not only on the blood of the host, but also on small arthropods inhabiting the host’s burrows). Most LM are closely related to diseases and belong to the category of medical gamasid mites. Some mites not only cause gamasid dermatitis, but also have the ability to transmit diseases between wild animals and humans, such as
Laelaps jettmari and
Haemolaelaps glasgowiare, which carry the haemorrhagic fever with renal syndrome (HFRS) virus [
6–
8].
Accurate identification of species is the basis for phylogenetic research. However, morphological taxonomic identification is difficult because morphological characters vary with age, developmental stage, environment, and other factors [
9]. Since the early 21st century, DNA barcoding technology has rapidly developed, providing opportunities for species identification and gradually becoming one of the main trends in mite taxonomy and molecular systematics [
10,
11]. The mitochondrial genome is the only extra-nuclear genetic information carrier in animals, and it has the characteristics of matrilineal inheritance, small molecular weight, high mutation rate, minimal recombination, and a fast evolutionary rate [
12,
13]. The mitochondrial cytochrome oxidase subunit I (
cox1) gene is the most conserved of the 3 genes encoding cytochrome oxidase. It has been shown to be suitable for addressing phylogenetic questions over a range of taxonomic levels [
14]. At present, arthropod ectoparasites of terrestrial hosts have been studied much less than those of helminths and ectoparasites of aquatic animals [
15]. Most scholars on LM have focused on the presence or description of species and the detection of pathogens. Thus, the development of molecular biology techniques has allowed the identification not only of morphologically determined species, but also of morphologically nearly identical species [
16].
The phylogenetic relationships among the LM remain unclear. In this study, we determined the cox1 gene sequences of 5 LM and analyzed morphological characteristic features.
MATERIALS AND METHODS
Ethics statement
All methods and procedures used in the capture rodent process were in accordance with the guidelines and regulations approved by the Animal Ethics Committees at Dali University. The approval ID is MECDU-201806-11.
Collection of mites
Mite specimens were obtained from the body surfaces of 5 rodent species (Niviventer fulvescens, Eothenomys miletus, Rattus tanezumi, Rattus nitidus, and Callosciurus erythraeus), in Li-jiang City, Yunnan Province, China, in August 2018. The obtained specimens were transported and stored at the Institute of Pathogens and Vectors, Dali University.
Morphological identification
Specimens of mites were mounted individually in Hoyer’s medium using light microscopy for morphological identification. Since the specimens were sufficiently cleared during the tissue lysis stage of DNA extraction, the typical clearing procedures were not necessary. Identification was based on the morphological characteristics of the mites in reference [
1].
Genomic DNA was extracted from individual mite using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, California, USA). The cox1 genes were all amplified by PCR (High Fidelity PCR system, Roche, Mannheim, Germany) with primers of the cox1 gene (Sense: 5′-GGAGGATTTGGAAATTGATTAGTTCC-3′; Anti sense: 5′-CCCGGTAAAATTAAAATATAAACTTC-3′). The PCR cycle conditions for the gene amplification were: 3 min at 94°C, over 40 cycles of 1 min at 94°C, 1 min at 52°C, 1 min at 72°C, and a final extension step of 10 min at 72°C. The PCR products were analyzed by electrophoresis on a 1% agarose gel. A paired approach was used to sequence PCR products (Thermo Fisher Scientific Genome Sequecing Facility, Guangzhou, China).
Data analysis
Raw sequences were assembled and aligned using the Geneious 11.1 software program (Biometer, Auckland, New Zealand) [
17]. The DAMBE [
18] software is based on genetic distance analyzed sequence base substitution saturation and the detection of phylogenetic signals. The DnaSP 5.0 [
19] software analyzed nonsynonymous (Ka) and synonymous (Ks) substitution ratios. Phylogenetic trees were constructed based on Bayesian in Mrbayes 3.2.5 [
20]. MrBayes model chosen a total of 1,000,000 generations were run, with sampling every 1,000 generations, while 4 Markov chains were run, discarding the first 25% of generations as burn-in. The constructed phylogenetic trees were viewed and edited using Figtree 1.4.4 [
21].
RESULTS
Morphological characteristics
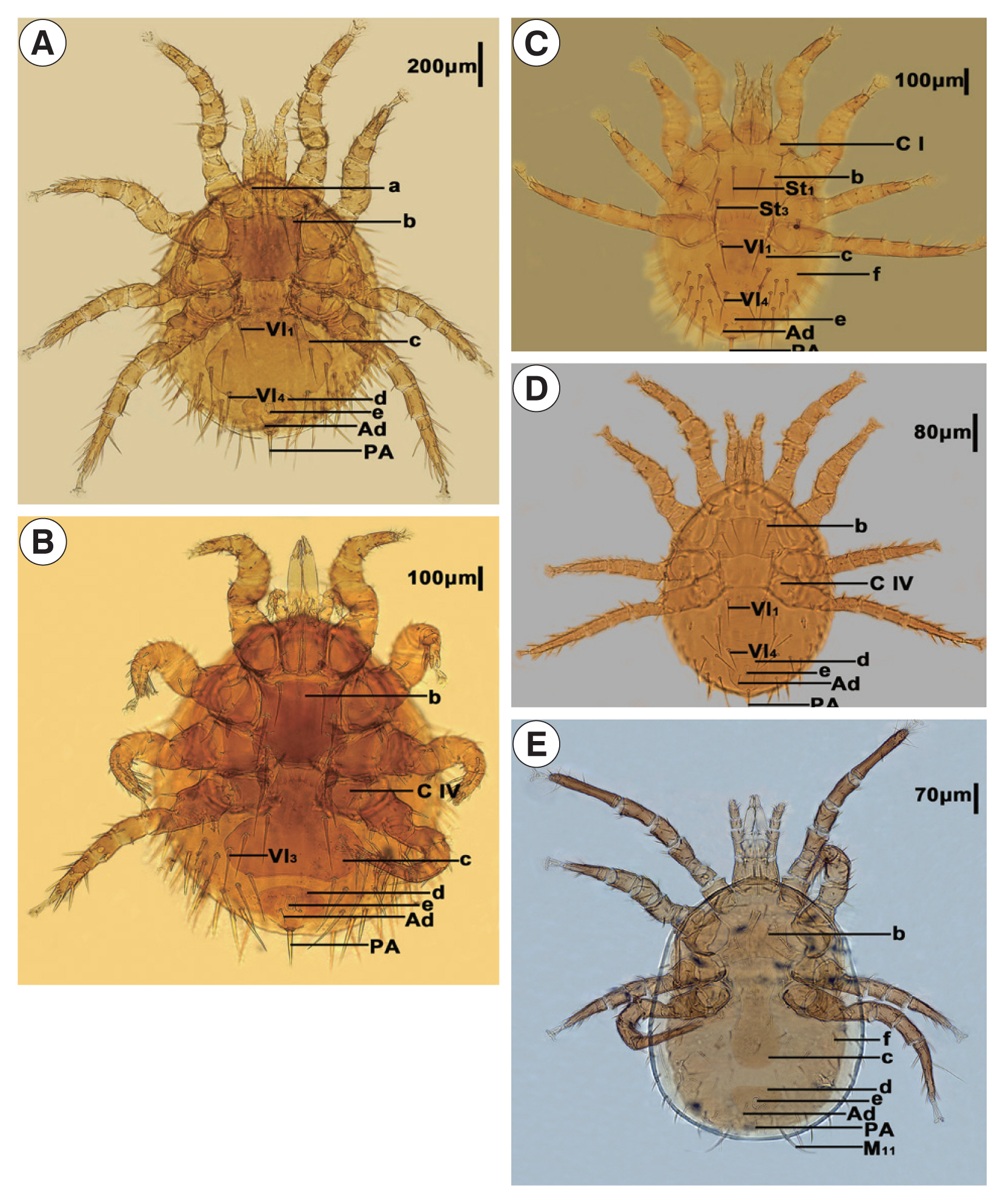
The 5 mites employed in this study were initially identified as
Laelaps echidninus (OL780835),
Laelaps nuttalli (OL810027),
Laelaps fukienensis (OL806574),
Laelaps chini (OL806586), and
Haemolaelaps traubi (OL810029). They are morphologically distinct (
Fig. 1,
Table 1).
The
cox1 gene lengths of 21 mites in 6 genera of Laelapinae, retrieved from GenBank Database (
www.ncbi.nlm.nih.gov/) ranged from 418 to 709 bp-long and from 61.0 to 74.6% in AT content (
Supplementary Table S1). The sequences of the 5 mites determined in this study ranged from 418 to 440 bp in length; the AT content ranged from 71.4 to 73.2%. The 5 mite sequences were aligned with other Laelapinae mites from GenBank and then trimmed to provide an equivalent sequence. There were 181 conserved sites, 230 variable sites, and 192 parsimony-informative sites, with no base insertions and deletions in the 411 bp alignment of
cox1 gene sequences. The non-synonymous mutation rate (Ka) and synonymous mutation rate (Ks) of
cox1 sequences were calculated using DnaSP 5.0 software, and the mean value of Ka/Ks was about 0.3.
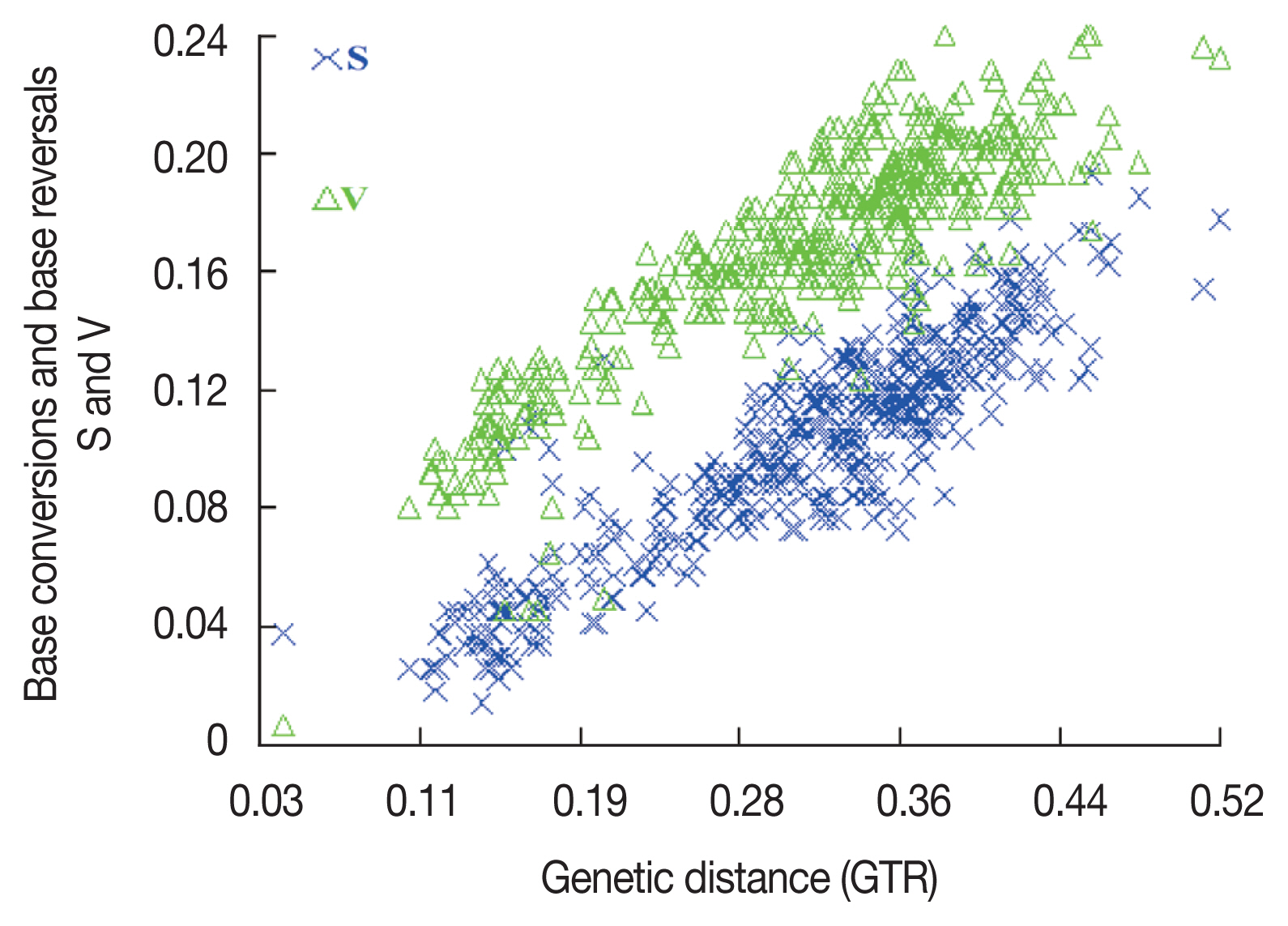
To ensure that all sequences were available for phylogenetic analysis, sequence base substitution saturation was analyzed using DAMBE software prior to constructing the phylogenetic tree. A scatter plot was constructed using genetic distance (GTR) as the horizontal coordinate and the number of base conversions and base reversals as the vertical coordinate (
Fig. 2). The number of base substitutions in the mt-DNA
cox1 gene sequence showed a significant linear relationship with the genetic distance, indicating that it can be used for phylogenetic analysis. Based on the mitochondrial
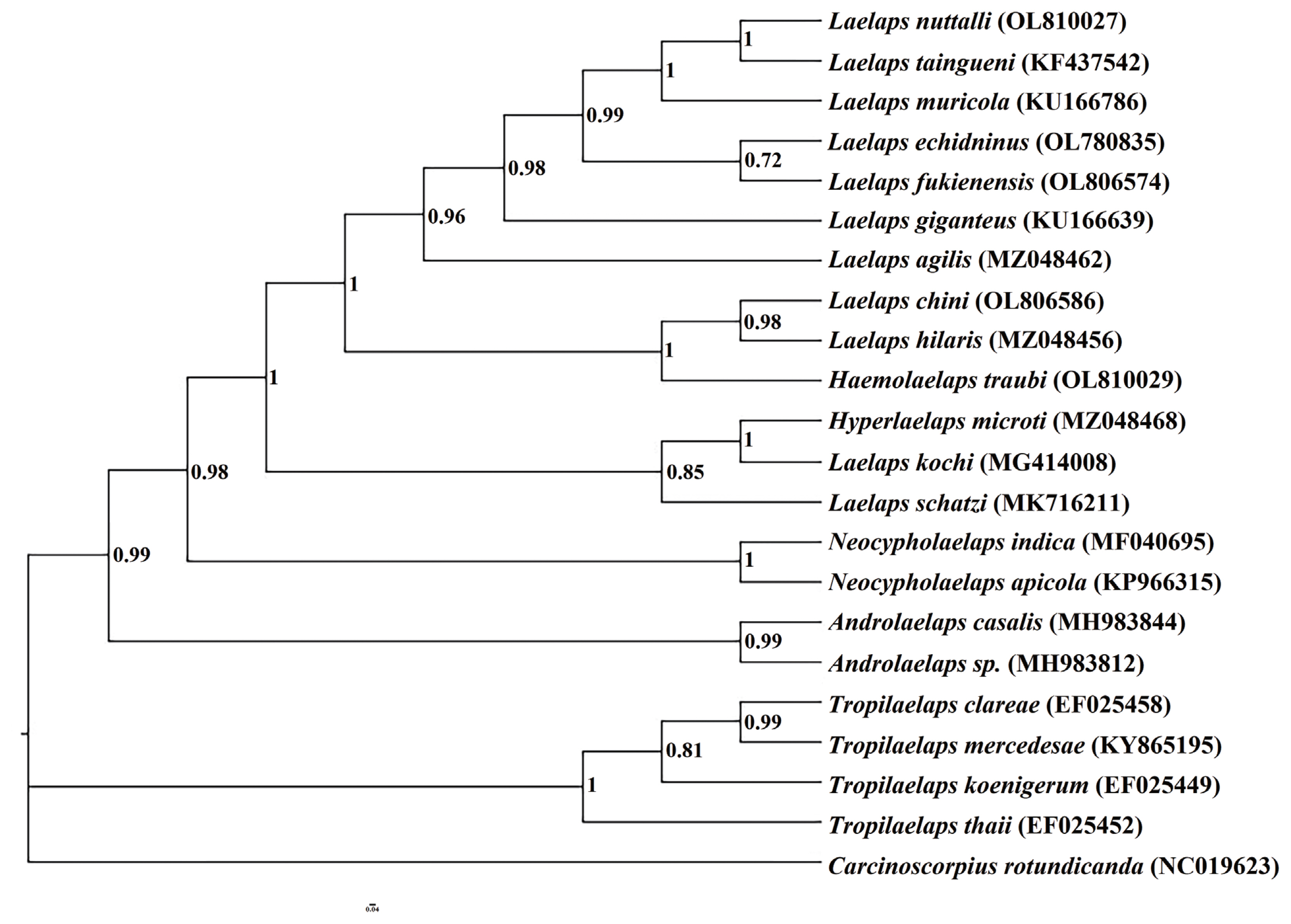
cox1 gene, the phylogenetic tree of Laelapinae mites was constructed by the BI method with
Carcinoscorpius rotundicauda (NC019623) as the outgroup (
Fig. 3). We observed that the Laelapinae is a monophyletic group, which is divided into 2 main branches (
Fig. 3). The first branch consists of 4 species of the genus
Tropilaelaps located at the base of phylogenetic tree, and the second branch contains the largest number of species, which consists of 17 species of 5 genera. The 5 species determined in this study are all included in the second branch.
Laelaps echidninus (OL780 835),
Laelaps nuttalli (OL810027),
Laelaps fukienensis (OL806 574) and
Laelaps chini (OL806586) are all clustered together with the species of the genus
Laelaps.
Haemolaelaps traubi (OL810029) does not form a separate clade, but clusters with 2 species (
Laelaps chini (OL806586) and
Laelaps hilaris (MZ048456)) of the genus
Laelaps. The phylogenetic tree constructed by the BI method can summarize the phylogenetic relationship of Laelapinae mites into (
Tropilaelaps + (
Neocycloplaelaps) + (
Laelaps +
Hyperlaelaps) + (
Haemolaelaps +
Laelaps)).
DISCUSSION
The morphological characters of 5 LM identified by light microscopy were found to be consistent with previous studies [
1]. The morphological characters of the 5 mites were distinctly different, and the main discriminatory character to distinguish the genus
Laelaps from the genus
Haemolaelaps was the number of setae on the genito-ventral plate. According to morphological observations,
Laelaps fukienensis (OL806574),
Laelaps chini (OL806586),
Laelaps echidninus (OL780835), and
Laelaps nuttalli (OL810027) conformed to the main morphological characters of the genus
Laelaps, i.e., the sternal plate has 3 pairs of setae, the genito-ventral plate has 4 pairs of setae, and the posterior part of the genito-ventral plate is obviously dilated.
Haemolaelaps traubi (OL810029) conforms to the main morphological characters of the genus
Haemolaelaps, i.e., the sternal plate is wider than long, the genito-ventral plate has only one pair of setae, and M
11 of
Haemolaelaps traubi (OL810 029) is particularly long as an important character for the identification of this species. Taxonomic identification of the mites is mainly based on traditional morphological characters [
11]. LMs are diverse, and many morphological characters are highly variable and indistinguishable in specific environments, while morphological similarities and ambiguous characters occur within the same genus. Therefore, the identification of LM by traditional morphological methods alone has some limitations. In order to get more objective results, we used both morphological and molecular evidence to identify 5 LM. This establishes a foundation for future research into the intraspecific phylogenetic relationships of LM.
In general, synonymous mutations are not subject to natural selection, whereas non synonymous mutations are subject to the action of natural selection. In evolutionary analysis, when Ka/Ks=1, natural selection is considered; when Ka/Ks <1, there might be a negative or purifying selection effect; and when Ka/Ks >1, it is considered that there is a positive or directional selection effect [
22]. In the present study, an average Ka/Ks ratio of 0.3 was found for 21 mites of Laelapinae, indicating that the Laelapinae mites experienced a purifying selection effect. This means that the
cox1 gene has evolved slowly and is highly conserved in the Laelapinae mites, making it suitable for exploring species taxonomy and phylogenetic relationships.
When we constructed the phylogenetic tree of Laelapinae employing the mitochondrial
cox1 gene sequences, we found that Laelapinae is a monophyletic group, with a relatively clear phylogenetic relationship. The species of the same genus always get together preferentially, with high node support rates, indicating that the intra-genus difference is less than the inter-genus difference. The genus
Tropilaelaps was located at the base of phylogenetic tree, showing that the genus
Tropilaelaps is an older taxon in the Laelapinae.
Laelaps fukienensis (OL806574),
Laelaps chini (OL806586),
Laelaps echidninus (OL780835), and
Laelaps nuttalli (OL810027) are well clustered with other species of the genus
Laelaps and have a high node support rates. This result indicates that the molecular taxonomy of these 4 species is completely consistent with the traditional morphological classification. Since the genera
Haemolaelaps and
Hyperlaelaps each involve only one species, namely
Haemolaelaps traubi (OL810029) and
Hyperlaelaps microti (MZ048468), they cluster with the species of the genus
Laelaps and do not form a separate cluster group. This observation differs from the existing taxonomic system of Laelapinae mites and requires further elaboration. The parasite depends primarily on the host for survival. Previous studies have reported that the phylogenetic relationship of the parasite is affected by the phylogenetic relationship of the parasite’s host [
23,
24]. Therefore,
Haemolaelaps traubi (OL810029) and
Hyperlaelaps microti (MZ048468) with 4 mites of the genus
Laelaps (
Laelaps chini (OL806586),
Laelaps hilaris (MZ048456),
Laelaps kochi (MG414008), and
Laelaps schatzi (MK716211)) clustered separately, which may be related to the relatively close kinship of the parasitic hosts of these species. Secondly, the genera
Haemolaelaps and
Hyperlaelaps may show similar morphological characteristics to the genus
Laelaps (for example, the dorsal plate setae are not very long or short, the chelicerae are thick, and the sternal plate has 3 pairs of setae) [
1]. In composing the phylogenetic tree, the species of the genera
Haemolaelaps and
Hyperlaelaps preferentially aggregate with the species of the genus
Laelaps. While the genus
Hyperlaelaps had been originally proposed as a subgenus of the genus
Laelaps [
25], many researches did not follow this conclusion. Close clustering of species from different genera has occurred in studies on the phylogenetic relationships of the
sesarmid crab, leading to the realignment of related species and the establishment of new genera [
26]. The phenomenon of close clustering of species of different genera (
Haemolaelaps and
Laelaps,
Hyperlaelaps and
Laelaps) has also been observed in Laelapinae. Based on this, identity of the species of the genera
Haemolaelaps and
Hyperlaelaps may need to be redefined.Can the genera
Hyperlaelaps and
Haemolaelaps be considered subgenera of the genus
Laelaps, or can
Haemolaelaps traubi (OL810029) and
Hyperlaelaps microti (MZ048468) be considered members of the genus
Laelaps? The validation of this hypothesis will require more in-depth and comprehensive morphological and molecular studies related to more species of the 2 genera.
At present, the phylogeny of LM has been relatively little studied, and the delineation of its taxonomic status and phylogenetic status need to be further refined. Due to the limited amount of information contained in the cox1 gene fragment utilized in this study, it may still be insufficient in resolving the affinities of some species, and the external morphological characters of some species are less different and still diverge in their taxonomic and evolutionary status. It is necessary to investigate more gene sequences as well as more species numbers to further define the taxonomic status and phylogenetic relationships of LM by combining morphological characters.
Notes
-
The authors declare that they have no conflicts of interests
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China [No. 32060143 and 31660314 to W Dong]. The data that support the findings of this study are available in the National Center for Biotechnology Information (NCBI) at [
https://www.ncbi.nlm.nih.gov/], reference number [OL806574, OL806586, OL780835, OL810027, OL810029]. We thank Ting Chen for assistance with an initial phylogenetic tree analysis
Fig. 1Morphological characteristics of Laelapinae spp. from China. (A) Laelaps echidinus, (B) Laelaps fukiensis, (C) Laelaps chini, (D) Laelaps nuttalli, and (E) Haemolaelaps traubi. a, Tritos ternum; b, Sternal plate; c, Genito-ventral plate; d, anal plate; e, anus; f, metapodal plate; C I, Coxa I; C IV, Coxa IV; Vl1, Genito-ventral plate first pair of setae; Vl3, Genito-ventral plate 3rd pair of setae; Vl4, Genito-ventral plate 4th pair of setae; St1, Sternal plate first pair of setae; St3, sternal plate 3rd pair of setae; Ad, adanal setae; PA, postanal setae.
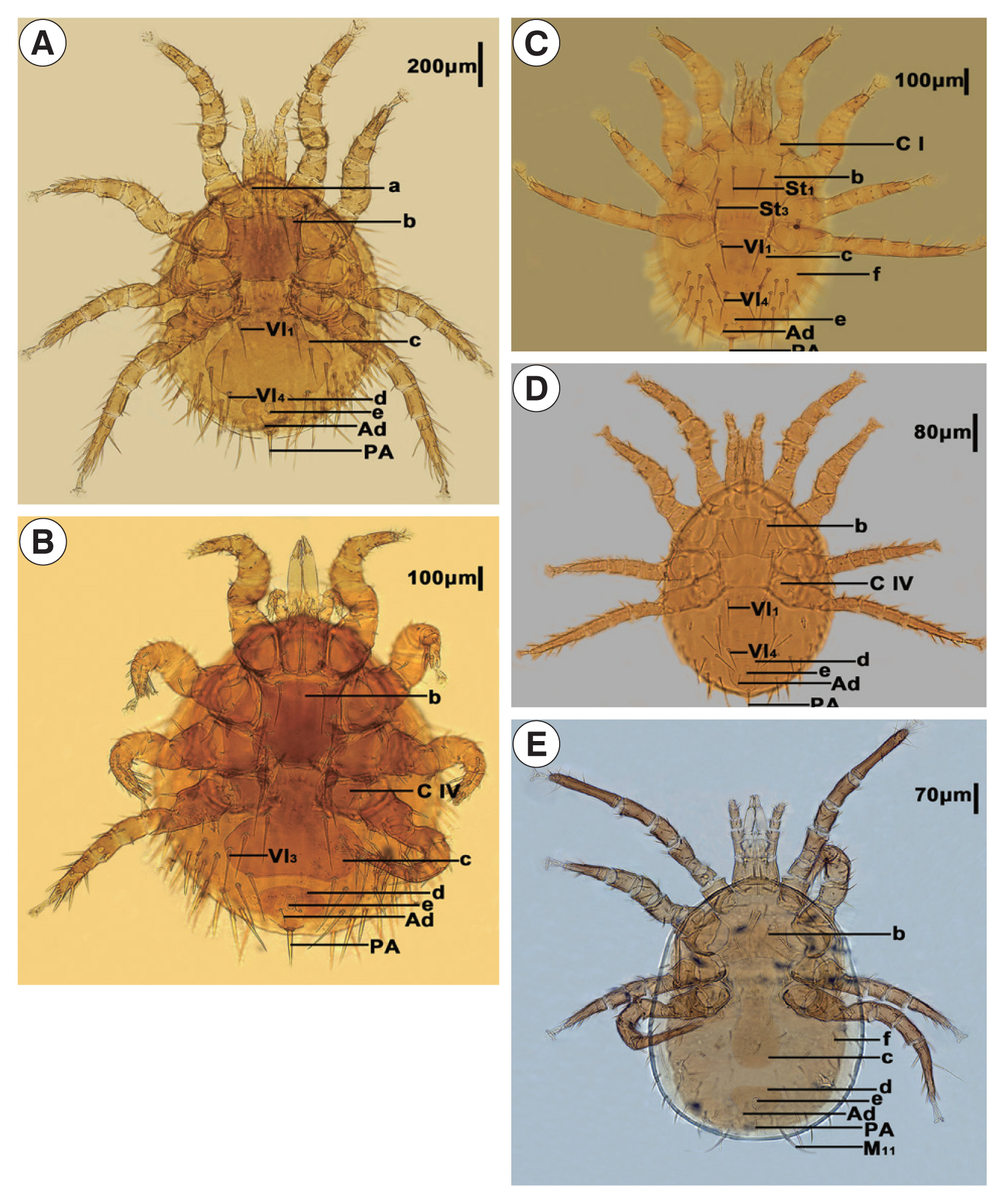
Fig. 2Saturation analysis of base substitutions.

Fig. 3Phylogenetic relationships of the laelapin mites based on cox1 gene.
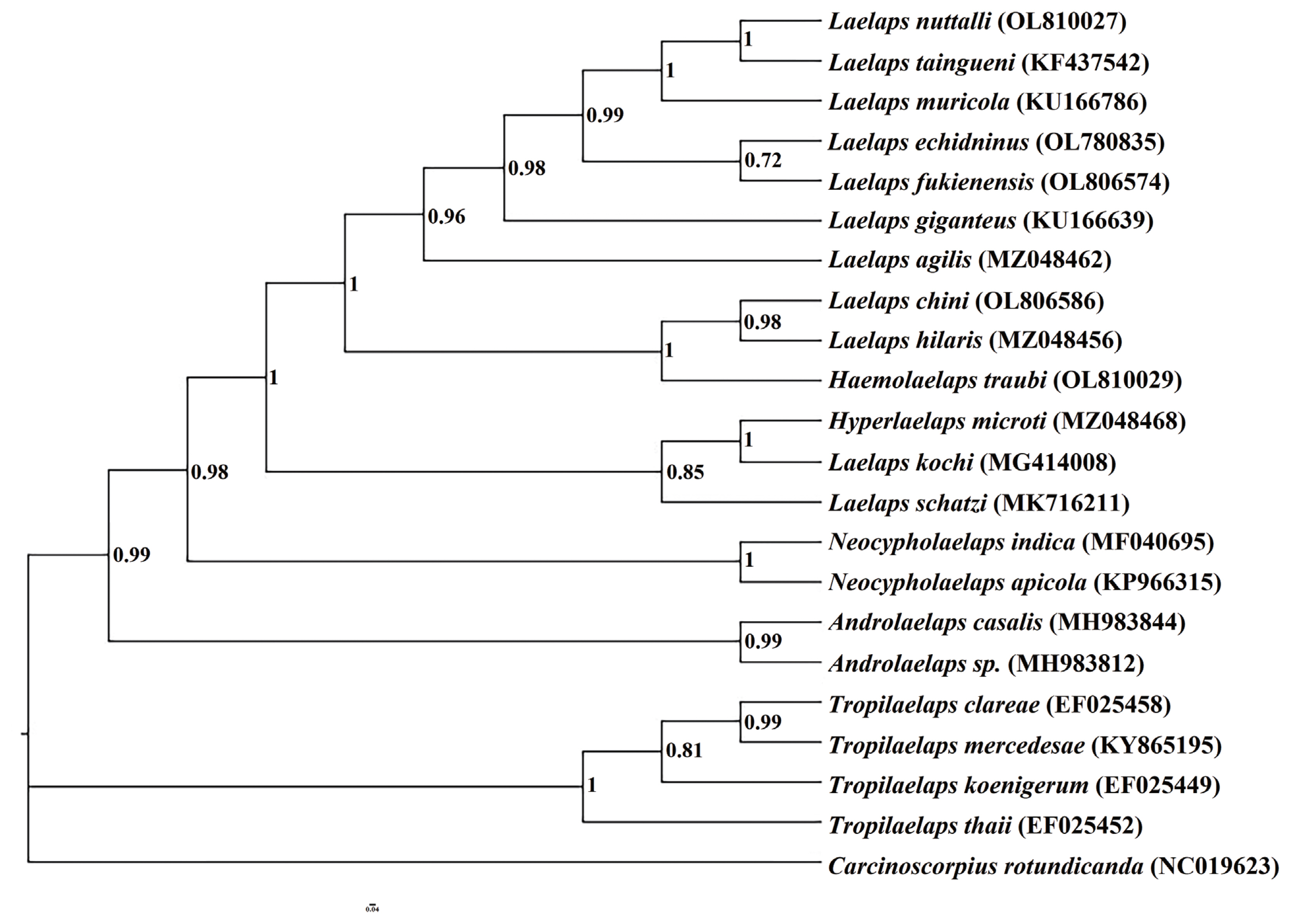
Table 1Identification basis of Laelapinae spp. from China
Table 1
|
Species |
Morphological characters |
|
Laelaps echidninus (OL780835) |
Length of the sternal plate is longer than width of it. Both sides of genital ventral plate are extremely expanded after Vl1, the trailing edge is deep and concave inward, whereas the spacing of Vl1 is less than Vl4. The distance between genito-ventral plate and anal plate is less than the length of anus, which shows a narrow groove. The front end of anal plate is wide and round, and the rear end is narrow and sharp. Adanal setae is located behind the rear end of anus, and the end reaches to the base of postanal setae. Tritosternum is far from the leading edge of sternal plate, and the space is much larger than the width of tritosternum. |
|
Laelaps fukienensis (OL806574) |
Length of the sternal plate is longer than width of it. The rear end of genito-ventral plate is wide and flat, with widest at Vl3 level, and extremely expanded after Coxa IV. The distance between genito-ventral plate and anal plate is slightly less than the length of anus. The front end of the anal plate is protruded slightly. Adanal setae is smaller, located at the level of the trailing edge of anus, and the end reaches to the base of postanal setae. Postanal setae is thick. |
|
Laelaps chini (OL806586) |
Length of the sternal plate is width than longer of it, the end of St1 can exceed the base of St3, and there is only one thick and short spiny hair on the trailing edge of Coxa I, whereas the other is an ordinary seta. Genito-ventral plate is nearly vase shaped, and the rear end is a little straight. The spacing of genito-ventral plate seta Vl1 is significantly greater than that of Vl4. Adanal setae is shorter than postanal setae, located at the level of the trailing edge of anus, and the end exceeds the base of postanal setae. Metapodal plate is in the shape of wheat grain. |
|
Laelaps nuttalli (OL810027) |
Length of the sternal plate is width than longer of it, the middle part of the leading edge is flat, and the trailing edge is concave. Genito-ventral plate is expanded after Coxa IV, the rear end is flat, and the space between Vl1 and Vl4 being almost equal. The anal plate is inverted pear shaped, the leading edge is relatively straight, and the adanal setae is located at the level of the trailing edge of anus, which does not reach to the base of postanal setae. |
|
Haemolaelaps traubi (OL810029) |
M11 is particularly long, which is one of the important characteristics of this species. The width of sternal plate is more than the length, and the middle part of the front edge is single peaked, but the boundary is not very obvious. The trailing edge is shallow and concave. Genito-ventral plate is long, the rear half is expanded slightly, the trailing edge is round and blunt, and there is only a pair of seta. The anal plate is a rounded triangle, and the adanal setae is about of the same length as that of the postanal setae, but the postanal setae is thicker than the adanal setae. Metapodal plate is in the shape of a short rod. |
References
- 1. Teng GF, Wang DQ, Gu YM, Meng YC. Economic Insects Fauna of China. Fasc 40 Acari: Demanyssoideae. Science Press; Beijing, China. 1993, pp 1-391 (in Chinese).
- 2. Lareschi M, Velazco PM. Laelapinae Mites (Acari: Parasitiformes: Laelapidae) Parasitic of Sigmodontine Rodents from Northern Peru, with the Description of a New Species from Akodon-aerosus (Rodentia: Cricetidae: Sigmodontinae). J Parasitol 2013;99:189-193. https://doi.org/10.1645/GE-3241.1
- 3. Radovsky FJ. The evolution of parasitism and the distribution of some dermanyssoid mites (Mesostigmata) on vertebrate hosts. In Houck MA ed, Mites: Ecological and Evolutionary Analyses of Life-History Patterns. Springer; New York, USA. 1994, pp 186-217.
- 4. Martins-Hatano F, Gettinger D, Bergallo HG. Ecology and host specificity of laelapine mites (Acari: Laelapidae) of small mammals in an Atlantic Forest area of Brazil. J Parasitol 2002;88:36-40. https://doi.org/10.2307/3285387
- 5. Korallo-Vinarskaya NP, Vinarski MV, Khokhlova IS, Shenbrot GI, Krasnov BR. Intraspecific variation of body size in a gamasid mite Laelaps clethrionomydis: environment, geography and host dependence. Parasitol Res 2015;114:3767-3774. https://doi.org/10.1007/s00436-015-4606-9
- 6. Wharton GW, Cross HF. Studies on the feeding habits of three species of laelaptid mites. J Parasitol 1957;43:45-50. https://doi.org/10.2307/3274753
- 7. Moro CV, Chauve C, Zenner L. Vectorial role of some dermanyssoid mites (Acari, Mesostigmata, Dermanyssoidea). Parasite 2005;12:99-109. https://doi.org/10.1051/parasite/2005122099
- 8. Krantz GW, Walter DE. A Manual of Acarology. 3rd ed. Texas Tech University Press; Lubbock, USA. 2009, pp 1-812.
- 9. Shen Y, Guan L, Wang D, Gan X. DNA barcoding and evaluation of genetic diversity in cyprinidae fish in the midstream of the Yangtze River. Ecol Evol 2016;6:2702-2713. https://doi.org/10.1002/ece3.2060
- 10. Hajibabaei M, Singer GA, Hebert PD, Hickey DA. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet 2007;23:167-172. https://doi.org/10.1016/j.tig.2007.02.001
- 11. Young MR, Moraza ML, Ueckermann E, Heylen D, Baardsen LF, Lima-Barbero JF, Gal S, Gavish-Regev E, Gottlieb Y, Roy L, Recht E, Adouzi ME, Palevsky E. Linking morphological and molecular taxonomy for the identification of poultry house, soil, and nest dwelling mites in the Western Palearctic. Scientific Reports 2019;9:5784-5791. https://doi.org/10.1038/s41598-019-41958-9
- 12. Boore JL. Animal mitochondrial genomes. Nucleic Acids Res 1999;27:1767-1780. https://doi.org/10.1093/nar/27.8.1767
- 13. Dong WG, Dong YL, Guo XG, Shao RF. Frequent tRNA gene translocation towards the boundaries with control regions contributes to the highly dynamic mitochondrial genome organization of the parasitic lice of mammals. BMC Genomics 2021;22:598-615. https://doi.org/10.1186/s12864-021-07859-w
- 14. Lin RQ, Qiu LL, Liu GH, Wu XY, Weng YB, Xie WQ, Hou J, Pan H, Yuan ZG, Zou FC, Hu M, Zhu XQ. Characterization of the complete mitochondrial genomes of five Eimeria species from domestic chickens. Gene 2011;408:28-33. https://doi.org/10.1016/j.gene.2011.03.004
- 15. Krasnov BR, Vinarski MV, Korallo-Vinarskaya NP, Khokhlova IS. Ecological correlates of body size in gamasid mites parasitic on small mammals: abundance and niche breadth. Ecography 2013;36:1042-1050. https://doi.org/10.1111/j.1600-0587.2012.00140.x
- 16. Zhou CJ, Feng MX, Tang YT, Yang CX, Meng XL, Nie GX. Species diversity of freshwater shrimp in Henan Province, China, based on morphological characters and COI mitochondrial gene. Ecol Evol 2021;11:10502-10514. https://doi.org/10.1002/ECE3.7855
- 17. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thiere T, Ashton B, Meintjes P, Durmmond A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647-1649. https://doi.org/10.1093/bioinformatics/bts199
- 18. Xia X. DAMBE 7: new and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol 2018;25:1550-1552. https://doi.org/10.1093/molbev/msy073
- 19. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009;25:1451-1452. https://doi.org/10.1093/bioinformatics/btp187
- 20. Ronquist F, Teslenko M, van der Mark PVD, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61:539-542. https://doi.org/10.1093/sysbio/sys029
- 21. Rambaut A. FigTree v1.4.4. [Internet]; Available from: http://tree.bio.ed.ac.uk/software/figtree/
- 22. Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet 2002;18:486-487. https://doi.org/10.1016/S0168-9525(02)02722-1
- 23. du Toit N, van Vuuren BJ, Matthee S, Matthee CA. Biome specificity of distinct genetic lineages within the four-striped mouse Rhabdomys pumilio (Rodentia: Muridae) from southern Africa with implications for taxonomy. Mol Phylogenet Evol 2012;65:75-86. https://doi.org/10.1016/j.ympev.2012.05.036
- 24. Engelbrecht A, Matthee S, du Toit N, Matthee CA. Limited dispersal in an ectoparasitic mite, Laelaps giganteus, contributes to significant phylogeographic congruence with the rodent host, Rhabdomys. Mol Ecol 2016;25:1006-1021. https://doi.org/10.1111/mec.13523
- 25. Zachvatkin AA. Organization of the genus Laelaps (Acarina, Parasitiformes) and the question of its epidemiological significance. Parazitologicheskii Sbornik 1948;10:50-75. (in Russian).
- 26. Shahdadi A, Schubart CD. Taxonomic review of Perisesarma (Decapoda: Brachyura: Sesarmidae) and closely related genera based on morphology and molecular phylogenetics: new classification, two new genera and the questionable phylogenetic value of the epibranchial tooth. Zool J Linn Soc 2018;182:517-548. https://doi.org/10.1093/zoolinnean/zlx032