Abstract
Phylogeny is the evolutionary history of a group or the lineage of organisms and is reconstructed based on morphological, molecular and other characteristics. The genealogical relationship of a group of taxa is often expressed as a phylogenetic tree. The difficulty in categorizing the phylogeny is mainly due to the existence of frequent homoplasies that deceive observers. At the present time, cladistic analysis is believed to be one of the most effective methods of reconstructing a phylogenetic tree. Excellent computer program software for phylogenetic analysis is available. As an example, cladistic analysis was applied for nematode genera of the family Acuariidae, and the phylogenetic tree formed was compared with the system used currently. Nematodes in the genera Nippostrongylus and Heligmonoides were also analyzed, and the validity of the reconstructed phylogenetic trees was observed from a zoogeographical point of view. Some of the theories of parasite evolution were briefly reviewed as well. Coevolution of parasites and humans was discussed with special reference to the evolutionary relationship between Enterobius and primates.
-
Key words: phylogeny, host-parasite relationship, zoogeography, nematoda, coevolution
Systems of nature: why do we classify?
All animals seem to have an instinctive ability to find suitable food and to avoid poisonous materials. Apparently, humans also possessed such ability from the beginning of the evolution, because it was necessary for them to be able to distinguish edible source of foods from the harmful ones for survival. Thus, the classification is an essential nature in humans. The ancient people empirically realized that there were rigid systems in nature presented by the movements of the sun, moon, constellations, and the cycle of the seasons. It was quite natural for our ancestors to conceive the idea that there must be a solitary system which living things obey. Studies on the natural system of living things is nothing more than discovering its structure and classifying them according to the similarities observed. Humans are animals with high sensitivities in visual senses, thus, classifying them by visible characters is a basic tendency. Hence, the classification is mainly based on the shape (= morphology).
An effort to find a system among the organisms was first expressed in 'Historia Animalium' by Aristotle in ancient Greece. The purpose of described philosophy (including natural sciences in those days) was to uncover the systems hidden in nature. The tradition has been maintained after the acceptance of Christianity. It was the mission of European scientists to prove the system created by Almighty God. Carl von Linné, the founder of modern systematics, established the methodology of classification in his 'Systema Naturae' (1735-1758), that has been adopted almost unchanged until today. In those days, the concept of evolution had not yet been developed. Hence, the systematics were so-called 'artificial classification' based on the resemblance in appearance which is quite different from the 'natural classification' that reflects the process of evolution precisely.
The idea of evolution occurred separately, but almost simultaneously, to Charles R. Darwin who made the expedition to the Galapagos Islands, and Alfred R. Wallace who surveyed animals in the Malay Archipelago (the area from the Malay Peninsula to New Guinea). The theory of evolution exerted a powerful influence on various fields of science and philosophy. In systematics, a tendency arose to make the system coincident with the process of evolution (= phylogeny). Ernst H. Haeckel took K.E. von Baer's law that "the earliest embryonic stages of related organisms are identical and distinguishing features develop later in ontogeny" from the viewpoint of evolution. He advocated the famous but incorrect law that "ontogeny repeats phylogeny". Haeckel also first presented a phylogenetic tree that reclassified organisms based on suggested evolutionary relationships. The phylogenetic tree modified by the 'neo-Haeckelian school' is still adopted in many textbooks. Phylogenetic trees have been presented not only for higher taxonomic groups but also for genera and species based on evidence in morphology, embryology, ecology, and other biological disciplines.
Schools in modern systematics
It is said that there are three major "schools" in modern systematics, i.e., evolutionary systematics, numerical taxonomy and cladistics. Evolutionary systematics, a descendant of traditional systematics, aims to reconstruct the phylogeny based on the degree of resemblance in observed characters. The premise of this systematics is that more phylogenetically distant ancestors and descendants may share fewer similarities than phylogenetically closer ancestors and descendants. However, is a phylogenetic tree based on such an observation is really correct? There are numerous homoplasies among highly diversified organisms. A homoplasy is a structural resemblance due to parallelism and convergent evolution rather than to common ancestry. Even if a well-experienced researcher, often an authority in the field, presents a phylogenetic tree, how can we verify its validity? The difficulty in reconstructing phylogeny is mainly due to frequent homoplasies.
Homoplasy of course exists frequently among parasites. Here I would like to share my experiences with a nematode of the family Acuariidae (Spirurida). Acuariid nematodes are parasitic in birds but rarely in mammals. In October 1992, we described a new acuariid,
Tikusnema javaense, in the new genus
Tikusnema (
Hasegawa et al., 1992). This nematode was collected from the alimentary canal of
Rattus argentiventer, a pest rodent in rice cultivation in Indonesia. Only one month later, British and Indonesian researchers published another paper describing a new acuariid,
Molinacuaria indonesiensis, from the same rat species in a nearby locality in Indonesia (
Gibbons et al., 1992). Both are apparently conspecific. Such cases are not uncommon in science, especially in the field of taxonomy. The problem is that we classified this nematode as a new genus in the subfamily Seuratiinae, whereas Gibbons et al. (
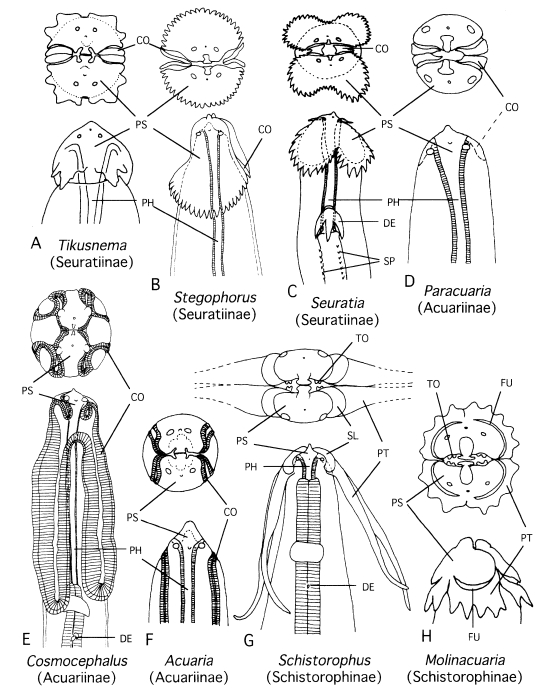
1992) placed it in the known genus of the subfamily Schistorophinae. The cause of these different classifications was the interpretation of small structures in the cephalic region. This nematode has four alate projections on the head (
Fig. 1 A). Gibbons et al. (
1992) may have considered them as ptilina that are specific to Schistorophinae (see
Wong and Lankester, 1985), whereas we judged that they were not homologous with ptilina but only with homoplasy. In the Schistorophinae, the ptilina are independent from the pseudolabia, and hence there is a furrow or suture between the pseudolabium and ptilinum (
Figs. 1 G, H). In the present nematode, the projections are continuous from the pseudolabia without an intervening furrow. Therefore, we believe that our interpretation is valid (cf.
Smales, 1995).
The above case may not be very complicated. However, nature is quite tricky, and it is often difficult to determine whether morphological characteristics are indeed homoplasy. How can we judge whether something is homoplasy or not? Experience is not enough to distinguish homoplasies. Thus, the phylogenetic trees presented by evolutionary systematists are more or less arbitrary even though they are expected to reflect evolutinary processes to certain extent. Numerical taxonomy is critical of the subjectivity of evolutionary systematics. The purpose of numerical taxonomy is to construct a strict classification system based on the distance between species calculated by multivariate analysis (such as cluster analysis) of numerical data obtained. Numerical taxonomy claims that taxonomy should be independent from the estimations of phylogeny. However, many taxonomists are not satisfied with such an artificial system and rather seek a system that reflects phylogeny.
How can we know the phylogeny? - Cladistic analysis
In order to move beyond the limit of traditional systematics, cladistics was established by the German entomologist, W. Hennig (
1966). The procedure of cladistic analysis can be summarized as follows. At first, each of the characters (morphological, embryological, ecological or sometimes distributional) of the OTU (operational taxonomic unit; species or group to be used for analysis) is classified as either plesiomorphic (i.e. ancestral) or apomorphic (i.e. derived). This process is called 'character argumentation' or 'polarization'. Then, '0' is assigned to plesiomorphic characters and '1' (or a higher number) to apomorphic characters in order to create a data matrix. Then, the 'tree length' (expressed as the number of steps in changes of the characters) is calculated for all of the possible tree forms, and the minimum length tree is selected as the most likely one (principle of parsimony) (
Eldridge and Cracraft, 1980;
Wiley, 1981).
Cladistic analysis is also applicable to molecular data such as the amino acid sequence of proteins and the base sequence of DNA. In these cases, the time of branching can be estimated based on the substitution rate of amino acids or bases. Recently, for phylogenetic analysis of molecular data, the neighbor-joining method and the maximum likelihood method have often been applied (
Saitou and Nei, 1987;
Nei, 1987). These methods are based on complicated statistical theories. However, analysis is easily carried out by using excellent computer program software such as PHYLIP, PAUP or MacClade (for information on software for phylogenetic analysis, visit the internet WWW site
http://evolution.genetics.washington.edu/phylip.html). Following a great deal of debate, cladistics seems to have become the leading field of systematics at the present time. Cladistical methodology is also frequently employed by evolutionary systematists to test their hypotheses on phylogeny.
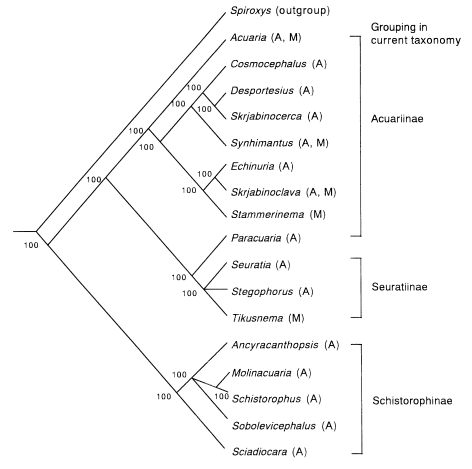
Now, let us try to make a phylogenetic tree through cladistic analysis. Materials are the genera of Acuariidae, including the nematode mentioned above. According to traditional taxonomy, the nematodes of Acuariidae are divided into three subfamilies, Acuariinae, Seuratiinae and Schistorophinae. Seventeen genera of which the morphology has been described in detail are subjected to analysis, i.e., nine genera in Acuariinae (
Acuaria,
Cosmocephalus,
Desportesius,
Echinuria,
Paracuaria,
Skrjabinocerca,
Skrjabinoclava,
Stammerinema,
Synhimantus), three genera in Seuratiinae (
Seuratia,
Stegophorus,
Tikusnema) and five genera in Schistorophinae (
Ancyracanthopsis,
Molinacuaria,
Schistor-ophus,
Sciadiocara,
Sobolevicephalus) (
Figs. 1 A-H). For a cladistic analysis, an outgroup is usually necessary, and here the genus
Spiroxys (Spirurina: Gnathostomatidae) has been chosen. Fifteen characters were selected, and the polarized results are shown in
Table 1. The data matrix based on these characters is shown in
Table 2. The cephalic structures (characters A to C and O) are significant characters of Acuariidae, and thus they are weighted twice as much as other characters.
These data were analyzed with the PAUP 4.0 program (
Swofford, 1998) and the two shortest trees with same topology were obtained by branch-and-bound search algorithm (
Fig. 2). Based on this tree, it is suggested that Schistorophinae first derived from the stem of ancestral Acuariidae, and then Acuariinae and Seuratinae were separated. As a whole, this tree seems congruent with the current classification by traditional taxonomy. However, the subfamily Acuariinae is not monophyletic because
Paracuaria belongs to another monophyletic group with Seuratiinae.
As shown in this example, the results by cladistic analysis often conflict with the classification systems presented by traditional or evolutionary systematists. The most famous case might be the results of the cladistic analysis of vertebrate groups based on morphological characters. According to these results, all of the reptilian groups, i.e., turtles, squamates, crocodilians are paraphyletic. From the cladistic viewpoint, a phylogenetically valid group should be monophyletic. Thus, the group traditionally known as Reptilia is not recognizable and should be grouped with birds to form a monophyletic 'Reptilia-Aves'. Such a grouping is far different than our usual concept, and hence elicited severe opposition and rejection from traditional and evolutionary systematists. Both the evolutionary systematic and cladistic analyses of morphological characters placed turtles near the base of the tree, whereas crocodiles were rather close to birds. More recently, however, cladistic analysis of DNA base sequence has revealed that turtles are much closer to crocodiles (
Hedges and Poling, 1999). An analysis using molecular data often gives quite different results from that based on morphological data.
Theories of parasite evolution
Since the 19th century, various theories have been proposed for the evolution of parasites. Even parasitologists today are predisposed to believe that parasites are 'lower' or 'inferior' organisms, wholly dependent on their hosts. In early the 20th century, H. Fahrenholz, a famous German specialist of sucking lice, considered that parasite speciation is strictly parallel to host speciation. According to his theory, the phylogenetic tree of parasites must be in the same form as that of their hosts, and hence each host species must have its own specific parasites. If a parasite seems to have wider host range, it merely means that its morphology or specificity has not been studied thoroughly. Such a theory was accepted until the 1960. Lothar Szidat, a trematode researcher from Argentine, considered that evolutionary trends which drive a host also drive its parasite. Harold W. Manter, a trematode researcher from the U.S.A., hypothesized that a parasite evolves more slowly than its host, and becomes to exhibit more specificity when the period of its association with the host is prolonged (see
Brooks and MacLennan, 1993, for review of these theories and historical backgrounds).
Meanwhile, Brooks & McLennan (
1993), leading evolutionary biologists studying parasite phylogeny, criticized these previous theories. They claimed that all of these theories were based on 'myths'. These myths included: (1) host specificity is unique and an important feature of parasite evolution; (2) parasites are simple and regenerative rather than free-living organisms; (3) parasites exhibit high adaptive plasticity. They, having proved that these 'myths' lack actual bases, stressed that evolution of parasites occurs in the same way as that of free-living organisms. Thus, their phylogeny should be reconstructed based on the characters of parasites themselves, not based on the host phylogeny.
Host-Parasite Coevolution
The term 'coevolution' was first coined to express the evolutionary relationship existing between plants and the insects feeding on them. Phenomena regarded as coevolutionary are observed in various examples such as pollinator insects and flowers, mimics and their models, and parasites and their hosts. The definition of coevolution could be summarized as: the interdependent evolution of two or more species having an obvious ecological relationship, often restricted to cases in which the interactions are beneficial to both species but is also used for any evolutionary interaction between species having some degree of interdependence, such as a parasite and its host' (
Lincoln et al., 1998). Typical coevolution occurs specifically, simultaneously and alternately between two organisms although there are many atypical cases. Futuyma and Slatkin (
1983) and Thompson (
1994) have published valuable textbooks on coevolution including host-parasite coevolution. Kim (
1985) edited an important book on the coevolution of parasitic arthropods and mammalian hosts. Durette-Desset (
1985) and Beveridge (
1986) published brilliant works on the coevolutionary relationships of helminth parasites and vertebrate hosts. Interesting discussions on parasite coevolution have also been presented by Brooks and McLennan (
1993).
Parasites do not always pledge allegiance to their hosts. If a parasite adapts to a new suitable host species, the possibility of evolutionary success increases. Sprent (
1968) proposed some interesting terms, i.e., 'heirloom parasites' for those that follow host evolution, and 'souvenir parasites' for those that adapt to hosts of a different phylogeny. Adaptation of a parasite to a new host that differs phylogenetically from its original one is called 'capture', 'host switching' or 'shift'.
Human parasites and coevolution
If parasites usually coevolve with their hosts, human parasites have also coevolved with primates. Probably, the most famous example is the pinworm, as shown in the study on coevolution of pinworms and primates by Brooks and Glen (
1982). Species of the genus
Enterobius have been recorded in various primates ranging from lemurs to humans. Interestingly, each primate species (or genus) harbors own
Enterobius species. Brooks and Glen (
1982) reconstructed the phylogenetic tree of
Enterobius by cladistic analysis, and proved that it closely resembles that of the host primates. Such a coincidence of host and parasite phylogenies suggests that the
Enterobius coevolved with the host primates rather strictly.
This 'strict' pinworm-primate coevolution apparently depends on the quite simple life history of the pinworm. The female pinworm lays eggs on the perianal skin of the host and the eggs soon become infective. Then they are ingested orally. Thus the life cycle is maintained in and on a host body. Such an exceptionally simple life history seems to provide less of an opportunity to acquire a new host than for other parasites that require a long period in an external environment or in intermediate hosts to become infective. Presumably, the host specificity of
Enterobius is not real but only seems so since there is little opportunity for the ingestion of pinworm eggs by other primate species. Indeed, chimpanzees under captivity are sometimes infected with the human pinworm,
E. vermicularis, instead of their specific pinworm,
E. anthropopitheci (
Hasegawa and Kinjo, 1996). Apparently, captive conditions enhance the opportunity to acquire infective ova from humans through contaminated food.
The pinworm-primate coevolution theory was stirred by the proposal of the 'two-species hypothesis' of human pinworms. Hugot (
1983) and Hugot and Toute-Schaefer (
1985) claimed that besides the human pinworm
E. vermicularis, one more species,
E. gregorii they called it, exists. Both species are distinguished from each other by the structure of the spicule and the pericloacal sclerotized plates in males, but females are indistinguishable. If there are two species of human pinworms, the process of their evolution should be reconsidered. Hugot (
1983) surmised that
E. vermicularis and
E. gregorii are sister species. If this is actually the case, the pinworms have speciated before the speciation of host humans. Meanwhile, Brooks and McLennan (
1994) suggested by cladistic analysis that the two pinworms are not sister species, and proposed the 'double-origin hypothesis': humans coevolved with
E. gregorii in Africa, and on their subsequent dispersal to Asia,
E. vermicularis was acquired from the hylobatid monkeys distributed there.
However, these arguments on evolutionary position of
E. vermicularis and
E. gregorii came to an unexpected end when the 'two' species were found to be synonymous. These 'two' species have been recorded in various localities around the world, often concurrently. Moreover, such concurrent infection was also seen in chimpanzees reared in a zoological garden (
Hasegawa and Kinjo, 1996). These ecological findings strongly suggest that these pinworms are synonyms. By examining numerous worms collected alive from a patient, it was proven that
E. gregorii is only a younger stage of
E. vermicularis, and that the spicule transformations from
E. gregorii-type to
E. vermicularis-type occur during growth (
Hasegawa et al., 1998). The specimens used by Hugot (
1983) to establish
E. gregorii were expelled by anthelmintics and were presumably much deformed, leading to wrong conclusion.
Careful consideration may be necessary in discussing the coevolution of parasites in such a host species as humans that has established a quite different ecological niche during recent evolution. Even if there is no natural definitive host other than humans, this does not mean that the parasite has coevolved with primates. For example,
Ascaris lumbricoides is a common parasite in humans. However, any
Ascaris infection is exceptional among non-human primates. Although infection has been recorded from chimpanzees, most of the cases were diagnosed under captive conditions, suggesting that the nematode infection was acquired after being captured. Even in chimpanzees,
Ascaris infection is quite rare in the wild (
File et al., 1976;
Hasegawa et al., 1983;
Landsoud-Soukate et al., 1995).
Humans are the only definitive host for
Taenia solium and
T. saginata. Primates are less carnivorous, and there has been no case of adult
Taenia infection in wild primates (
Yamashita, 1963;
Myers and Kuntz, 1972;
Healy and Myers, 1973). Probably
Ascaris and
Taenia were acquired when our ancestors left arboreal life for open land habitats suitable to erect bipedalism, and began to interact with the ancestors of domestic animals or change their feeding habits. Thus, the coevolutionary history of these human parasites should be relatively shorter.
Zoogeography and phylogeny of parasites
The idea of evolution occurred almost simultaneously to Darwin and Wallace as mentioned above. Wallace is less famous than Darwin but his contribution to the establishment of the idea of evolution was not inferior to Darwin. His name has been retained in the Wallace's line, a famous zoogeographical boundary passing between Kalimantan (Borneo) and Sulawesi. Mammalian fauna differs greatly on either side of this line. For example, murid fauna of Sulawesi is composed of nearly 50 species in 14 genera of which 10 genera are endemic (
Musser and Holden, 1991). Except for several species that have extended their distribution as a result of human movements, there is no species common with any in Kalimantan. Murid rodents apparently originated on the Asian continent, and some ancestral murids managed to reach Sulawesi, New Guinea and Australia across the sea on quite rare occasions, and then evolved into various endemic species. Their parasites also apparently crossed the sea with the hosts, and thus this history must be reflected on the parasite fauna.
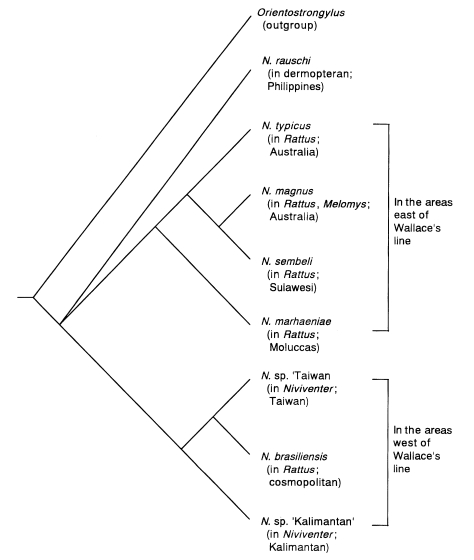
Let us examine the phylogeny of nematodes of the genus
Nippostrongylus (Trichostrongyloidea: Heligmonellidae) as an example. In this genus, besides the famous cosmopolitan
N. brasiliensis, parasitic in
Rattus rattus and
Rattus norvegicus, nine representatives have been found: eight species in endemic rats (including two species in Australia, one species each in the Moluccas, Sulawesi, Kalimantan, Taiwan and continental China), and one species in the Philippine dermopteran (
Hasegawa and Syafruddin, 1995;
Hasegawa and Tarore, 1995) (
Fig. 3). Eight of these species of which the morphology is relatively well known are analyzed here. As an outgroup,
Orientostrongylus tenorai (Trichostrongyloidea: Heligmonellidae) is employed. The morphological characters of these species are divided into plesiomorphic and apomorphic as stated in
Table 3. The data matrix formed is shown in
Table 4. This matrix was analyzed with the PAUP 4.0 program, and only one shortest tree was obtained by branch-and-bound search algorithm as shown in
Fig. 4. This tree suggests that the
Nippostrongylus species in endemic murids distributed in the areas east and west of the Wallace's line form monophyletic groups respectively. Presumably, the
Nippostrongylus lineage in east of the Wallace's line separated from the lineage of those west of the Wallace's line at an early stage of the evolution of
Nippostrongylus. As shown by this analysis, the zoogeographical boundary is reflected in the parasite fauna.
Relationship of parasite faunas between Korea and Japan
The sea between Korea and Japan also represents a zoogeographical boundary. In order to demonstrate whether this boundary is also recognizable in parasite fauna, let us examine the genus
Heligmonoides (Trichostrongyloidea: Heligmonellidae). The field mouse,
Apodemus agrarius, is famous as a reservoir of Hantaan virus in Korea. This small rodent is found in Korea, including Cheju Island, but is also distributed over a wide geographical range, including Taiwan, and from continental China to Europe. East Asian populations of
A. agrarius harbor an undescribed nematode of the genus
Heligmonoides (referred to here as
Heligmonoides sp. 'agrarius') (
Asakawa and Tenora, 1996). In Japan,
A. agrarius is not found, except in Senkaku Islands where this mouse is distributed and harbors
H. sp. 'agrarius' (
Hasegawa et al., 1993). Tsushima Island is very close geographically to the Korean Peninsula, but
A. agrarius is not found there. Instead, two species of
Apodemus, i.e.
A. speciosus and
A. argenteus, are distributed on the island. They do not host
H. sp. 'agrarius', but instead harbor
H. speciosus, a common parasite of
Apodemus spp. on the Japanese mainland and its surrounding islands (
Asakawa et al., 1991;
Asakawa, 1995). Apparently, the Korean Strait has been a rather strict barrier for the dispersal of the host and its parasites.
Interestingly,
Apodemus semotus in the Taiwan highlands harbors another species,
Heligmonoides taiwanensis (
Hasegawa, 1990). This means that different
Apodemus species have specific
Heligmonoides. Besides these representatives,
Heligmonoides has been also recorded in other murid genera: four species in
Mus spp. in areas from Japan to Africa; four in
Maxomys spp. in Malaysia and Sulawesi, and several species in the endemic murines in the Ryukyu Archipelago and Taiwan (
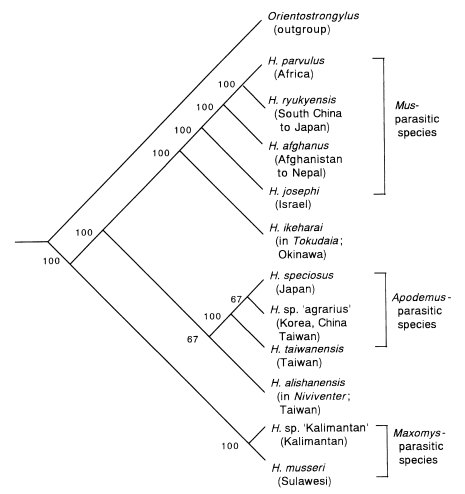
Hasegawa, 1991). Cladistic analysis has been applied to their morphological characters (
Fig. 5 A-D) and the data matrix shown in
Tables 5 and
6, respectively. As an outgroup,
Orientostrongylus tenorai is again employed. By branch-and-bound search algorithm, three shortest trees were obtained, and the concensus tree of them is presented in
Fig. 6. The
Maxomys-parasitic species are shown to divert from other members at the most basal node. These species have many morphological characters inconsistent with other members of
Heligmonoides (
Fig. 5 D). This result by cladistic analysis may support the creation of the new genus
Maxomystrongylus for them by Hasegawa and Syafruddin (
1997). It is also notable that
Mus-parasitic and
Apodemus-parasitic species form distinct lineages. This may mean that ancestors of
Mus-parasitic and
Apodemus-parasitic species were divided at an early stage of
Heligmonoides evolution, and coevolved with their host genera separately.
How to reconstruct more reliable phylogenetic trees
Recent advances in the technology of molecular biology has made it easy to analyze DNA base sequences. As mentioned already, a phylogenetic tree reconstructed from molecular data often differs from that based on morphological data. By analysis using DNA base sequences, the time of branching can be estimated, and the results often coincide with the geological evidence. Some researchers, especially molecular systematists, claim that it is outdated to reconstruct a phylogenetic tree based on morphological data because subjectivity or arbitariness are frequently involved in morphological observation and character argumentation. However, homoplasies might also exist in DNA base sequences, and the base substitution rate has been proven to differ greatly by portions in DNA. Moreover, the DNA base sequences hitherto elucidated are of quite limited portions. The codes that are responsible for morphological characters are undoubtedly coded in the DNA sequence, but the process of expression of such specific characters is still in a black box.
It is an essential ability of humans to distinguish a species with their visual sense, and this species recognition by morphological observation will continue. Hence, morphology is an important base for any phylogenetic analysis. I believe that reconstruction of phylogeny based only on DNA molecule or only on morphology may lead to erroneous interpretation. Because we cannot go back to the past, phylogeny can be reconstructed only by approximation. A more reliable phylogenetic tree is expected to be obtained by critical analysis repeatedly using newer and better data.
Notes
-
This review was presented as an invited lecture for commenoration of the 40th anniversary of the Korean Society for Parasitology, 29 October 1999.
ACKNOWLEDGEMENTS
I wish to thank Dr. S. Y. Kang, President, and Dr. S. T. Hong, Secretary for Academic Affairs, the Korean Society for Parasitology, for their kindness in giving me this valuable opportunity to present my studies on parasite systematics. I am also indebted to Dr. H. Ota for his valuable suggestions, and Dr. S. Kamegai, Dr. M. Asakawa, Dr. K. Koyasu and Dr. S. Shiraishi, for their kindness in supplying relevant literature and/or nematode specimens, and to Prof. G. B. Rodgers for his kindness in reviewing the manuscript.
References
- 1. Asakawa M. A biogeographical study of the parasitic nematodes of Japanese Microtinae and Muridae with systematic and phylogenetic studies of the genera Heligmosomoides and Heligmosomum (Nematoda: Heligmosomidae). J Rakuno Gakuen Univ 1995;19:285-379.
- 2. Asakawa M, Tenora F. A check list of epidemiology of nematode parasites of the genus Apodemus (Murinae: Rodentia) throughout the world excluding Japan. J Rakuno Gakuen Univ 1996;20:181-213.
- 3. Asakawa M, Yamaugchi S, Fujino R, Ohbayashi M, Hasegawa H. Study on the helminth fauna of the Japanese wood and field mice, Apodemus spp., on the Tsushima and Iki Islands. Bull Biogeogr Soc Jpn 1991;46:59-68. (in Japanese with English summary).
- 4. Beveridge I. In Stone AR, Hawksworth DL eds, Coevolutionary relationships of the helminth parasites of Australian marsupials. Coevolution and Systematics. 1986, Oxford, UK. Clarendon Press; pp 93-117.
- 5. Brooks DR, Glen DR. Pinworms and primates: a case study in coevolution. Proc Helminthol Soc Wash 1982;49:76-85.
- 6. Brooks DR, MacLennan DA. Parascript - Parasites and the Language of Evolution. 1993, Washington, USA. Smithsonian Institution Press; p 429.
- 7. Durette-Desset MC. Le genre Nippostrongylus Lane, 1923 (Nematode-Heligmosomatid. Ann Parasitol Hum Comp 1970;45:815-821.
- 8. Durette-Desset MC. Trichostrongyloid nematodes and their vertebrate hosts: reconstruction of the phylogeny of a parasite group. Adv Parasitol 1985;24:239-306.
- 9. Eldridge N, Cracraft J. Phylogenetic Patterns and the Evolutionary Process: Method and Theory in Comparative Biology. 1980, New York, USA. Columbia University Press; p 349.
- 10. File SK, McGrew WC, Tutin CEG. The intestinal parasites of a community of feral chimpanzees, Pan troglodytes schweinfurthii. J Parasitol 1976;62:259-261.
- 11. Futuyma DJ, Slatkin M eds, Coevolution. 1983, Sunderland, Massachusetts, USA. Sinauer Association Inc.; p 555.
- 12. Gibbons LM, Grawshaw MT, Rumpus AE. Molinacuaria indonesiensis n. sp. (Nematoda, Acuarioidea) from Rattus argentiventer in Indonesia. Syst Parasitol 1992;23:175-181.
- 13. Hasegawa H. Nematodes of the family Heligmonellidae (Trichostrongyloidea) collected from rodents of the Ryukyu Archipelago and Taiwan. J Parasitol 1990;76:470-480.
- 14. Hasegawa H, Arai S, Shiraishi S. Nematodes collected from rodents on Uotsuri Island, Okinawa, Japan. J Helminthol Soc Wash 1993;60:39-47.
- 15. Hasegawa H, Kano T, Mulavwa M. A parasitological survey on the feces of Pygmy chimpanzees, Pan paniscus, at Wamba, Zaire. Primates 1983;24:419-423.
- 16. Hasegawa H, Kinjo T. Human pinworms collected from a chimpanzee, Pan troglodytes, in a zoo of Okinawa, Japan. J Helminthol Soc Wash 1996;63:272-275.
- 17. Hasegawa H, Shiraishi S, Rochman . Tikusnema javaense n. gen., n. sp. (Nematoda: Acuarioidea) and other nematodes from Rattus argentiventer collected in West Java, Indonesia. J Parasitol 1992;78:800-804.
- 18. Hasegawa H, Syafruddin . Hasanuddinia maxomyos n. gen. n. sp. and Heligmonoides musseri n. sp. (Nematoda: Heligmonellidae) collected from endemic murines of Sulawesi, Indonesia. J Parasitol 1994;80:781-788.
- 19. Hasegawa H, Syafruddin . Nippostrongylus marhaeniae sp. n. and other nematodes collected from Rattus cf. morotaiensis in North Halmahera, Molucca Islands, Indonesia. J Helminthol Soc Wash 1995;62:111-116.
- 20. Hasegawa H, Syafruddin . Maxomystrongylus yasumai gen. and sp. n. (Nematoda: Trichostrongylina: Heligmonellidae) collected from murid rodents in Kalimantan. J Helminthol Soc Wash 1997;64:263-268.
- 21. Hasegawa H, Takao Y, Nakao M, Fukuma T, Tsuruta O, Ide K. Is Enterobius gregorii Hugot, 1983 (Nematoda: Oxyuridae) a distinct species? J Parasitol 1998;84:131-134.
- 22. Hasegawa H, Tarore D. Nippostrongylus sembeli, new species (Nematoda: Heligmonellidae) collected from Rattus xanthurus of North Sulawesi, Indonesia. Raffles Bull Zool 1995;43:337-342.
- 23. Healy GR, Myers BJ. Intestinal helminths. Chimpanzee 1973;6:265-296.
- 24. Hedges SB, Poling LL. A molecular phylogeny of reptiles. Science 1999;283:998-1001.
- 25. Hennig W. Davis DD, Zangerl R, translators. Phylogenetic Systematics. 1966, Urbana, USA. University of Illinois Press; p 263.
- 26. Hugot JP. Enterobius gregorii (Oxyuridae, Nematoda), un nouveau parasite humain. Ann Parasitol Hum Comp 1983;58:403-404.
- 27. Hugot JP, Tourte-Schaefer C. Etude morphologique des deux oxyures parasites de l'homme: Enterobius vermicularis et E. gregorii. Ann Parasitol Hum Comp 1985;60:57-64.
- 28. Inglis WG. The nematodes parasitic in the gizzard of birds: a study in morphological convergence. J Helminthol 1965;39:207-224.
- 29. Kim KC ed, Coevolution of Parasitic Arthropods and Mammals. 1985, New York, USA. John Wiley & Sons; p 800.
- 30. Landsoud-Soukate J, Tutin CEG, Fernandez M. Intestinal parasites of sympatric gorillas and chimpanzees in the Lope Reserve, Gabon. Ann Trop Med Parasitol 1995;89:73-79.
- 31. Lincoln R, Boxshall G, Clark P. A Dictionary of Ecology, Evolution and Systematics. 1998, Cambridge, UK. Cambridge University Press; p 361.
- 32. Mawson PM. Trichostrongyles from rodents in Queensland, with comments on the genus Longistriata (Nematoda: Heligmosomatidae). Aust J Zool 1961;9:791-826.
- 33. Musser GG, Holden ME. Sulawesi rodents (Muridae: Murinae): Morphological and geographical boundaries of species in the Rattus hoffmanni group and a new species from Pulau Peleng. Bull Am Mus Nat Hist 1991;206:322-413.
- 34. Myers BJ, Kuntz RE. A checklist of parasites and commensals reported for the chimpanzee (Pan). Primates 1972;13:433-471.
- 35. Nei M. Molecular Evolutionary Genetics. 1987, New York, USA. Columbia University Press; p 512.
- 36. Quentin JC, Seureau S, Gabrion C. Cycle biologique d Acuaria anthuris (Rudolphi, 1819), nematode parasite de la pie. Z Parasitenk 1972;39:103-126.
- 37. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-425.
- 38. Smales LR. A review of the genus Tikusnema (Nematoda: Acuarioidea) with the description of a new species from the false water-rat, Xeromys myoides from Queensland. Trans Roy Soc South Aust 1995;119:89-94.
- 39. Sprent JFA. In Jackson GD ed, Evolutionary aspects of immunity in zooparasitic infections. Immunnity to parasitic animals. 1969, Vol. 1:New York, USA. Appleton-Century-Crofts; pp 3-62.
- 40. Swofford DL. PAUP (Phylogenetic analysis using parsimony). 1998, 4.0 (Beta Version). Sunderland, Massachusetts, USA. Sinauer Associates, Inc..
- 41. Tenora F. Parasitic nematodes of certain rodents from Afghanistan. Vest Cesk Spol Zool 1969;33:174-192.
- 42. Thompson JN. The Coevolutionary Process. 1994, Chicago, USA. University of Chicago Press; p 376.
- 43. Wiley EO. Phylogenetics: the Theory and Practice of Phylogenetic Systematics. 1981, New York, USA. John Wiley & Sons; p 439.
- 44. Wong PL, Lankester MW. Revision of the genus Schistorophus Railliet, 1916 (Nematoda: Acuarioidea). Can J Zool 1984;62:2527-2540.
- 45. Wong PL, Lankester MW. Revision of the genus Ancyracanthopsis Diesing, 1861 and description if a new genus Molinacuaria n. gen. (Nematoda: Acuarioidea). Can J Zool 1985;63:1556-1564.
- 46. Yamashita J. Ecological relationships between parasites and primates. I. Helminth parasites and primates. Primates 1963;4:1-96.
Fig. 1Semidiagrammatic illustrations of cephalic portions of nematodes in some acuariid genera.
A. Tikusnema;
B. Stegophorus;
C. Seuratia;
D. Paracuaria;
E. Cosmocephalus;
F. Acuaria;
G. Schistorophus; and
H. Molinacuaria. Subfamilies assigned in current taxonomy are given in the parentheses. Modified after
Inglis, 1965 (for
Seuratia),
Quentin et al., 1972 (
Acuaria),
Wong and Lankester, 1984 &
1985 (
Schistorophus,
Molinacuaria)
Hasegawa et al., 1992 (
Tikusnema). Abbreviations: CO, cordon; DE, deirid; FU, furrow; PH, pharynx; PS, pseudolabium; PT, ptilinum; SL, sublabium; SP, spine rows; TO, tooth.
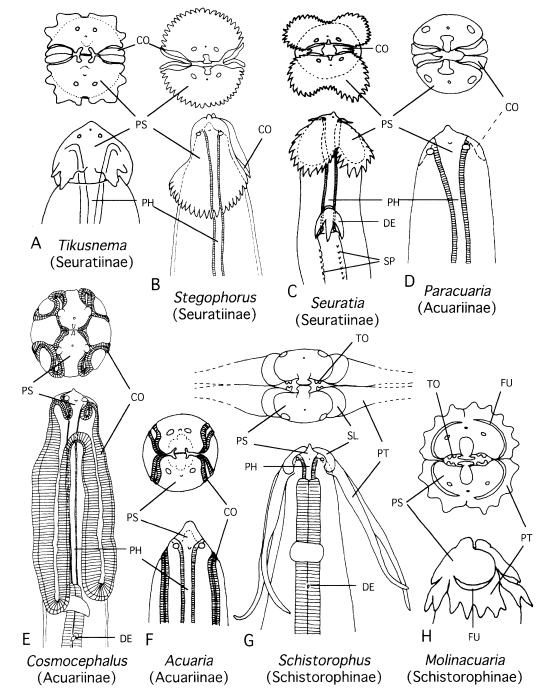
Fig. 2A consensus cladogram of the genera in Acuariidae computed by majority rule on 2 maximum parsimony trees obtained by branch-and-bound search analysis of the data in
Table 2. CI (consistency index) = 0.60. Numbers in the figure represent group frequencies (%).
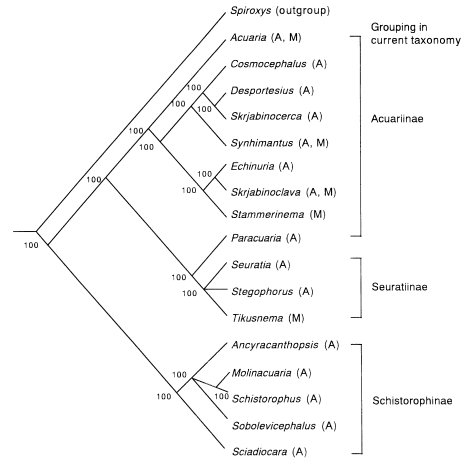
Fig. 3
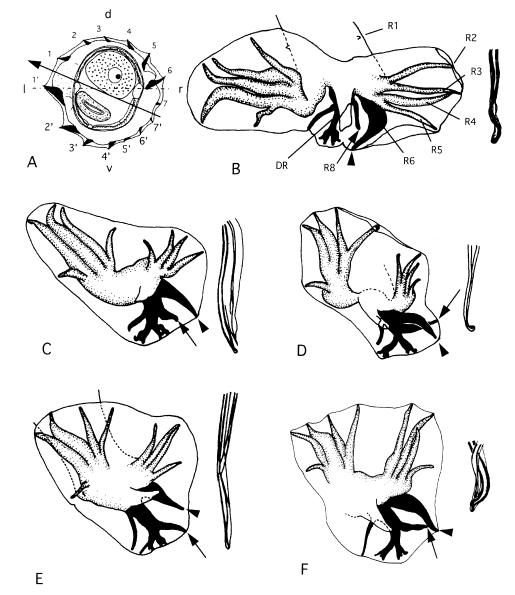
Fig. 4The maximum parsimony tree of
Nippostrongylus obtained by branch-and-bound search analysis of the data in
Table 4. CI = 0.73.
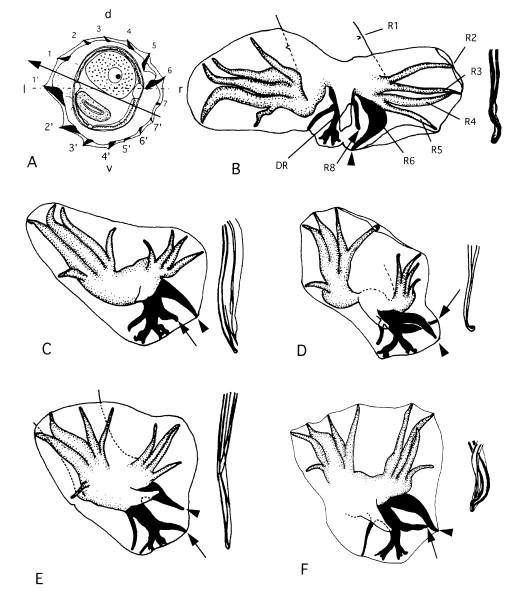
Fig. 5Morphological characteristics of several
Heligmonoides species.
A, B. Heligmonoides sp. 'agrarius' from
Apodemus agrarius of Korea; cross section in midbody (A) and bursa copulatrix, ventral view (B).
C. Cross section in midbody of
Heligmonoides afghanus from
Mus sp. of Nepal.
D. Cross section in midbody of
Heligmonoides musseri (=
Maxomystrongylus musseri) from
Maxomys musschenbroekii of Sulawesi (modified after
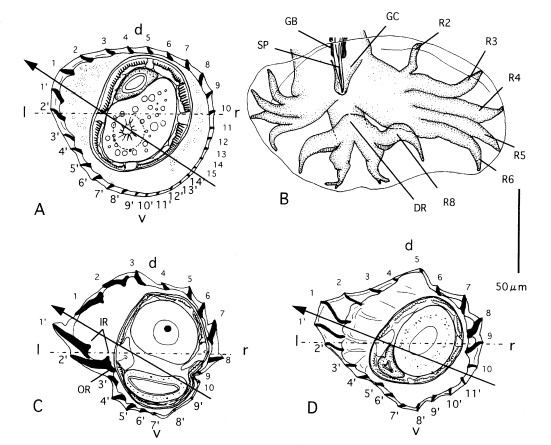
Hasegawa and Syafruddin, 1994). Horizontal line and arrowed line represent frontal axis and axis of orientation of ridges, respectively. Abbreviations: d, dorsal; DR, dorsal ray; GB, gubernaculum; GC, genital cone; l, left; IR, inner root of ridges at left lateral side; OR, outer root of ridges at left lateral side; r, right; R2-R8, ray 2-ray 8; SP, spicule; v, ventral.
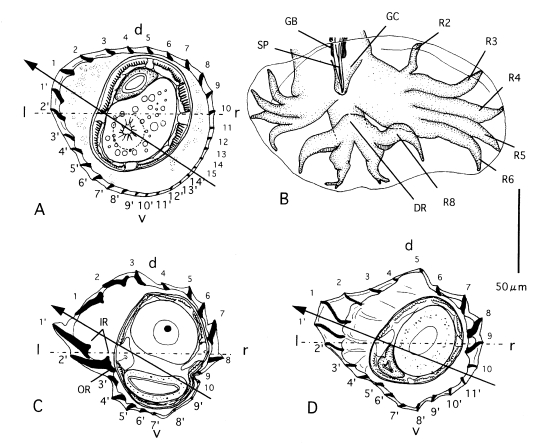
Fig. 6A consensus cladogram of the genus
Heligmonoides computed by majority rule on 3 maximum parsimony trees obtained by branch-and-bound search analysis of the data in
Table 6. CI = 0.57. Numbers in the figure represent group frequencies (%).
Table 1.Character analysis of Acuariidae
Table 1.
|
Characters |
Plesiomorphy (0) |
Apomorphy (1) |
Apomorphy (2) |
Apomorphy (3) |
Apomorphy (4) |
|
A. Cordon |
absent |
rudimentary |
straight, extending posteriorly |
recurrent and/or anastomose |
forming sinuosities |
|
B. Sublabia |
absent |
present |
|
|
|
|
C. Ptilina |
absent |
simple or weakly modified |
elongated or strongly modified |
|
|
|
D. transverse striation of pharynx |
present |
absent |
|
|
|
|
E. Deirids |
minute, spinose |
large, modified |
|
|
|
|
F. Body spines |
absent |
present |
|
|
|
|
G. Lateral alae |
absent |
present |
|
|
|
|
H. Caudal alae |
short |
long |
|
|
|
|
I. Pedunculate caudal papillae |
4 pairs |
6 pairs |
> 8 pairs |
|
|
|
J. Spicules |
quite dissimilar |
equal or subequal |
|
|
|
|
K. Position of vulva in body |
middle 1/3 |
posterior 2/3 |
near anus |
|
|
|
L. Uterus |
didelphic |
monodelphic |
|
|
|
|
M. Posterior margin of pseudolabium |
not serrated |
serrated |
|
|
|
|
N. Area rugosa in male |
absent |
few lines |
much modified |
|
|
|
O. Oral teeth |
absent |
present |
|
|
|
Table 2.Character matrix used in the reconstruction of the phylogenic relationship of the Acuariidae
Table 2.
|
Characters |
Plesiomorphy (0) |
Apomorphy (1) |
Apomorphy (2) |
Apomorphy (3) |
Apomorphy (4) |
|
A. Cordon |
absent |
rudimentary |
straight, extending posteriorly |
recurrent and/or anastomose |
forming sinuosities |
|
B. Sublabia |
absent |
present |
|
|
|
|
C. Ptilina |
absent |
simple or weakly modified |
elongated or strongly modified |
|
|
|
D. transverse striation of pharynx |
present |
absent |
|
|
|
|
E. Deirids |
minute, spinose |
large, modified |
|
|
|
|
F. Body spines |
absent |
present |
|
|
|
|
G. Lateral alae |
absent |
present |
|
|
|
|
H. Caudal alae |
short |
long |
|
|
|
|
I. Pedunculate caudal papillae |
4 pairs |
6 pairs |
> 8 pairs |
|
|
|
J. Spicules |
quite dissimilar |
equal or subequal |
|
|
|
|
K. Position of vulva in body |
middle 1/3 |
posterior 2/3 |
near anus |
|
|
|
L. Uterus |
didelphic |
monodelphic |
|
|
|
|
M. Posterior margin of pseudolabium |
not serrated |
serrated |
|
|
|
|
N. Area rugosa in male |
absent |
few lines |
much modified |
|
|
|
O. Oral teeth |
absent |
present |
|
|
|
Table 2.Character matrix used in the reconstruction of the phylogenic relationship of the Acuariidae
Table 2.
|
Genus |
Transformation seriesa)
|
|
Ab)
|
Bb)
|
Cb)
|
D |
E |
F |
G |
H |
I |
J |
K |
L |
M |
N |
Ob)
|
|
Acuaria
|
2 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
|
Ancyracanthopsis
|
0 |
1 |
2 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
|
Cosmocephalus
|
4 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Desportesius
|
3 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
0 |
2 |
1 |
0 |
0 |
0 |
|
Echinuria
|
3 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
|
Molinacuaria
|
0 |
0 |
2 |
0 |
0 |
0 |
0 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
1 |
|
Paracuaria
|
1 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Schistorophus
|
0 |
1 |
2 |
0 |
0 |
0 |
0 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
1 |
|
Sciadiocara
|
0 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
|
Seuratia
|
1 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
|
Skrjabinocerca
|
2 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
|
Skrjabinoclava
|
4 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
2 |
1 |
0 |
2 |
0 |
|
Sobolevicephalus
|
0 |
1 |
2 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
|
Stammerinema
|
3 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
|
Stegophorus
|
1 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
|
Synhimantus
|
3 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
|
Tikusanema
|
1 |
0 |
0 |
1 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
2 |
0 |
|
Spiroxysc)
|
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
Table 3.Character analysis of Nippostrongylus
Table 3.
|
Characters |
Plesiomorphy (0) |
Apomorphy (1) |
Apomorphy (2) |
|
A. No. of synlophe ridges in midbody |
> 15 |
< 15 |
|
|
B. Small accessory ridge |
present |
absent |
|
|
C. Ventro-right ridges |
present |
absent |
|
|
D. Size of bursal lobes |
right ≦ left |
right > left |
|
|
E. Distance between papillae 3-4 : 4-5 in right lobe |
3-4 ≧ 4-5 |
3-4 < 4-5 |
|
|
F. Length of rays 3 and 4 in right lobe |
3 << 4 |
3 = 4 |
|
|
G. Length of rays 3 and 4 in left lobe |
3 ≦ 4 |
3 > 4 |
|
|
H. Thickness of rays 5 and 6 in left lobe |
5 ≧ 6 |
5 < 6 |
|
|
I. Distance between papillae 4-5 : 5-6 in left lobe |
4-5 5-6/2 |
4-5 > 5-6/2 |
|
|
J. Rays 6 and 8 in left lobe |
separated |
close |
crossed |
|
K. Thickness of ray 8 in both lobes |
left = right |
left > right |
|
|
L. Level of ray 8 in both lobes |
same |
different |
|
|
M. Bifurcation shape of dorsal ray |
inverted ‘V’ |
inverted ‘U’ |
|
|
N. Twist of spicule tips |
absent or weak |
strong |
hooked |
|
O. Postvulval body |
conical |
constricted |
invaginated |
|
P. Gubernaculum |
present |
absent |
|
Table 4.Character matrix used in the reconstruction of the phylogenic relationship of Nippostrongylus
Table 4.
|
Species |
Transformation seriesa)
|
|
A |
B |
C |
D |
E |
F |
G |
H |
I |
J |
K |
L |
M |
N |
O |
P |
|
N. rauschi
|
1 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
|
N. typicus
|
1 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
1 |
0 |
2 |
1 |
0 |
|
N. magnus
|
1 |
1 |
0 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
2 |
1 |
0 |
|
N. sembeli
|
1 |
1 |
0 |
1 |
0 |
1 |
1 |
1 |
0 |
2 |
1 |
1 |
0 |
2 |
1 |
0 |
|
N. sp. ‘Taiwan’ |
1 |
1 |
0 |
1 |
1 |
0 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
|
N. sp. ‘Kalimantan’ |
1 |
1 |
0 |
1 |
1 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
|
N. brasiliensis
|
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
2 |
0 |
|
N. marhaeniae
|
1 |
1 |
1 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
1 |
0 |
|
Orientostrongylusb)
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
Table 5.Character analysis of Heligmonoides
Table 5.
|
Characters |
Plesiomorphy (0) |
Apomorphy (1) |
Apomorphy (2) |
|
A. No. of synlophe ridges in midbody |
< 15 |
15-25 |
> 25 |
|
B. Left lateral slender ridges |
absent |
present |
|
|
C. Longest left lateral ridge/body width |
< 1/10 |
1/10-1/5 |
> 1/5 |
|
D. Inclination of axis of orientation of ridges |
45° |
50-65° |
> 65° |
|
E. Length of spicules/worm length |
< 20% |
20-40% |
> 40% |
|
F. Genital cone protrusion |
strong |
slight |
flat |
|
G. level of dorsal ray bifurcation |
distal 1/3 |
middle 1/3 |
basal 1/3 |
|
H. Origin of ray 8 from dorsal ray bifurcation |
basal to |
same level |
distal to |
|
I. Shape of distal ends of spicules |
slender |
plow-like |
branched |
|
J. Ray 8 width (8): dorsal ray width (d) |
d > 8 |
d = 8 |
d < 8 |
|
K. Ray 3 width (3): ray 4 width (4) |
3 = 4 |
3 < 4 < 3 × 2 |
4 < 3 × 2 |
|
L. Distance between papillae 2-3 : 3-4 in left lobe of bursa |
2-3 > 3-4 |
2-3 = 3-4 |
2-3 < 3-4 |
|
M. Distance between papillae 4-5 : 5-6 in left lobe |
4-5 > 5-6 |
4-5 = 5-6 |
4-5 < 5-6 |
|
N. Distance between papillae 2-4 : 3-4 in right lobe |
2-3 > 3-4 |
2-3 = 3-4 |
2-3 < 3-4 |
|
O. Large ridge in female posterior body |
absent |
present |
|
|
P. Lateral diverticulum of vagina vera |
absent |
present |
|
|
Q. Size ratio of bursal lobes |
right < left |
right << left |
|
|
R. Posterior invagination in female |
absent |
present |
|
|
S. Inner root of left developed ridge |
absent or weak |
strong |
|
|
T. Outer root of left developed ridge |
absent or weak |
strong |
|
|
U. Wide space between left ventral ridges |
absent |
present |
|
Table 6.Character matrix used in the reconstruction of the phylogenic relationship of Heligmonoides
Table 6.
|
Species |
Transformation seriesa)
|
|
A |
B |
C |
D |
E |
F |
G |
H |
I |
J |
K |
L |
M |
N |
O |
P |
Q |
R |
S |
T |
U |
|
H. parvulus
|
1 |
0 |
2 |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
1 |
0 |
1 |
|
H. josephi
|
1 |
0 |
2 |
1 |
0 |
0 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
|
H. afghanus
|
0 |
0 |
2 |
1 |
0 |
0 |
2 |
2 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
1 |
0 |
1 |
1 |
1 |
|
H. ryukyensis
|
0 |
0 |
1 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
0 |
1 |
|
H. sp. ‘Kalimantan’ |
0 |
1 |
2 |
2 |
0 |
2 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
|
H. musseri
|
1 |
1 |
2 |
2 |
1 |
2 |
1 |
0 |
1 |
0 |
1 |
1 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
|
H. speciosus
|
2 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
|
H. taiwanensis
|
1 |
0 |
1 |
1 |
0 |
1 |
1 |
1 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
|
H. sp. ‘agrarius’ |
2 |
0 |
0 |
1 |
0 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
|
H. ikeharai
|
1 |
0 |
1 |
1 |
2 |
1 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
|
H. alishanensis
|
1 |
0 |
1 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
|
Orientstrongylusb)
|
0 |
0 |
0 |
0 |
0 |
2 |
1 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |