Abstract
The karyotypes of Biomphalaria tenagophila collected from Rio de Janeiro, Brazil were studied using the air-drying method. Somatic cells of this species had 2n=36. The 18 chromosome pairs were identified and classified into 3 groups. The diploid cell has 7 pairs of metacentric, 8 pairs of submetacentric, and 3 pairs of subtelocentric chromosomes. Observed chromosomes ranged from 2.4 to 6.4 µm, and the total length was 122.3 µm. This is the first report on the chromosome of B. tenagophila.
-
Key words: Biomphalaria tenagophila, karyotype, freshwater snail, Brazil
Biomphalaria tenagophila is a species of air-breathing freshwater snail, an aquatic pulmonate gastropod mollusk in the family Planorbidae.
B. tenagophila is a neotropical species, and its native distribution include Peru, Uruguay, Argentina, and southern Brazil [
1,
2,
3,
4]. Santos et al. [
5] was the first to record
B. tenagophila on Ilha Grande, Rio de Janeiro, in Brazil. This species is a medically important pest because of transferring the disease, intestinal schistosomiasis [
6]. Intestinal schistosomiasis is the most widespread of all types of schistosomiasis [
7,
8].
B. tenagophila is an intermediate host and a vector of
Schistosoma mansoni [
9].
S. mansoni infects about 83.31 million people worldwide [
10].
The study of an organism genome at the chromosomal level can be used in differentiating one species from another, i.e., the analysis of chromosome numbers, size, and centromere positions [
11,
12,
13]. In fact, modern cytogenetic techniques have since mid-1990s been adopted for studies of Gastropoda. The subfamily Biomphalarinae has been known to be a conservative group in regard to haploid chromosome numbers 18 (
Table 1). In this study, the karyotype of
B. tenagophila was studied in order to analyze their genetic relationships.
The 9 specimens used in this study were collected in Museu de Arte Moderna (22°54'48.12"S, 43°10'19.17"W), Rio de Janeiro, Brazil, September 2012, and examined shortly after collection (
Fig. 1). The chromosome preparations were made on gonad of the specimens by the usual air-drying method [
14]. The prepared slides were observed under an Olympus (BX51) microscope. The identical specimens used for this study have been deposited at the Department of Environmental Medical Biology, Kwandong University College of Medicine, Korea.
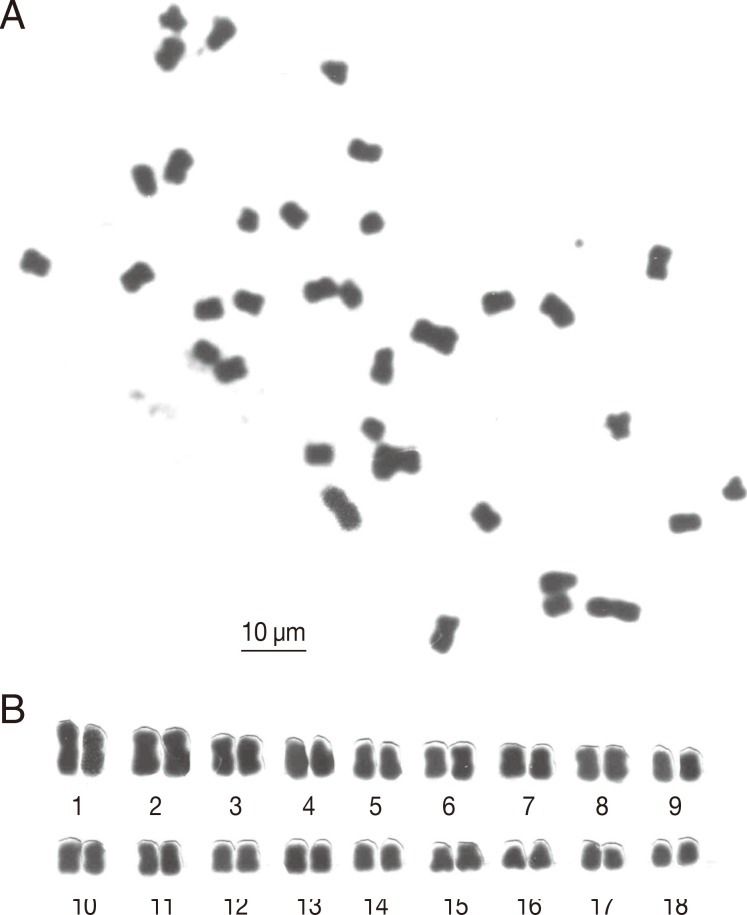
A microphotograph of somatic metaphase chromosomes and the karyogram are shown in
Fig. 2, and measurements of the chromosomes are presented in
Table 2.
Fig. 2B shows the karyotype of this species arranged by size. The present cytological preparations showed 10 well-spread mitotic cells on a slide. This species showed a diploid chromosome number of 2n=36, with 7 metacentric, 8 submetacentric, and 3 subtelocentric chromosome pairs. Observed chromosomes ranged from 2.4 to 6.4 µm in length. This species was no inter-specimen variability in chromosome counts. Also, sexual dimorphism of chromosomes was not found in this study.
Most species of the
Biomphalaria have been investigated and at present,
B. glabrata,
B. pfeifferi,
B. madagascariensis,
B. sudanica,
B. tanganyicensis,
B. alexandrina, and
B. truncatus have a haploid chromosome complement of 18 (
Table 1). The chromosomes are relatively small and monomorphic. Conservation of chromosome number has been pointed out for many gastropoda groups [
15]. Burch [
16], in the 1960s, utilized preparations from the ovo-testis and reported a haploid complement of 18; however, these results were deducted from the meiotic stage, where it was often different to identify each and every chromosome. However, with the development of cytological methods such as hypotonic treatment of tissue samples and pre-treatment with colchicines (air drying technique), clearer deductions were possible on the chromosome number and morphology [
11]. The morphology of mitotic metaphase chromosomes has been reported in 10 species of the subfamily Biomphalrinae with n=18 and 2n=36 (
Table 1).
The present results revealed that the diploid chromosome number of
B. tenagophila is 36. With regard to karyotype analysis in the genus
Biomphalaria, only 4 paper about
B. glabrata,
B. truncates, and
B. alexandria has been published [
17,
18,
19]. Though the chromosome number is the same between
B. glabrata,
B. alexandria,
B. truncates, and
B. tenagophila, the karyotype is different.
B. glabrata and
B. alexandria have 10 pairs of metacentric, 4 pairs of submetacentric, 2 pairs of acrocentric, and 2 pairs of telocentric chromosomes, and
B. truncatus has 10 metacentric, 4 submetacentric, 2 subtelocentric, and 2 telocentric chromosomes, whereas
B. tenagophila has 7 metacentric, 8 submetacentric, and 3 subtelocentric chromosomes. The results of this study on the mitotic metaphase chromosomes provided a much more elucidated observation of the chromosomes of this species. Further studies are needed with various analyses for the investigation of the karyo-systematic evolution, and accumulation of karyological information is very much required in the subfamily Biomphalarinae.
Notes
-
We have no conflict of interest related to this study.
References
- 1. Paraense WL, Corrêa LR. Differential susceptibility of Biomphalaria tenagophila populations to infection with a strain of Schistosoma mansoni. J Parasitol 1978;64:822-826.
- 2. da Silva CL, Soares MS, Barreto MGM. Occurrence of Biomphalaria tenagophila and Disappearance of Biomphalaria straminea in Paracambi, RJ, Brazil. Mem Inst Oswaldo Cruz 1997;92:37-38.
- 3. Majoros G, Fehér Z, Deli T, Földvári G. Establishment of Biomphalaria tenagophila snails in Europe. Emerg Infect Dis 2008;14:1812-1814.
- 4. Scholte RGC, Carvalho OS, Malone JB, Utzinger J, Vounatsou P. Spatial distribution of Biomphalaria spp., the intermediate host snails of Schistosoma mansoni, in Brazil. Geospatial Health 2012;6:S95-S101.
- 5. dos Santos SB, Miyahira C, de Lacerda LEM. First record of Melanoides tuberculatus (Müller, 1774) and Biomphalaria tenagophila (d'Orbigny, 1835) on Ilha Grande, Rio de Janeiro, Brazil. Biota Neotropica 2007;7:361-364.
- 6. Pointier JP, Pointier JP, David P, Jarne P. Biological invasions: the case of planorbid snails. J Helminthol 2005;79:249-256.
- 7. Fitzpatrick JM, Protasio AV, Mcardle AJ, Williams GA, Johnston DA, Hoffmann KF. Use of genomic DNA as an indirect reference for identifying gender-associated transcripts in morphologically identical, but chromosomally distinct, Schistosoma mansoni cercariae. PLoS Negl Trop Dis 2008;2:e323.
- 8. Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis 2006;6:411-425.
- 9. Borda CE, Rea MJF. "Biomphalaria tenagophila potencial vector of Schistosoma mansoni in the Paraná River basin (Argentina and Paraguay)". Mem Inst Oswaldo Cruz 2007;102:191-195.
- 10. Crompton DW. How much human helminthiasis is there in the world? J Parasitol 1999;85:397-403.
- 11. Thiriot-Quiévreux C. Advances in chromosomal studies of gastropod mollusks. J Mol Stu 2003;69:187-201.
- 12. Hamta A, Adamovic T, Samuelson E, Helou K, Behboudi A, Levan G. Chromosome ideograms of the laboratory rat (Rattus norvegicus) based on high-resolution banding, and anchoring of the cytogenetic map to the DNA sequence by FISH in sample chromosomes. Cytogenet Genome Res 2006;115:158-168.
- 13. Odoemelam EC. Genomic analysis of the fresh water mollusk Biomphalaria glabrata to understand host: parasite interactions. Brunel University, UK; 2009, p 242 Ph.D. thesis.
- 14. Park GM. Karyotypes of Korean endemic land snail, Koreanohadra koreana (Gastropoda: Bradybaenidae). Korean J Malacol 2011;27:87-90.
- 15. Patterson CM. Chromosomes of molluscs. Proc Symp Moll II. 1968; India: Mar Biol Ass; 1969. 635-686.
- 16. Burch JB. Chromosome numbers and systematic in euthyneuran snails. Proc First Eur Malacol Cong. 1962; 1965. 215-241.
- 17. Raghunathan L. The karyotype of Biomphalaria glabrata, the snail vector of Schistosoma mansoni. Malacologia 1976;15:447-450.
- 18. Bakry FA, Garhy MFEl. Comparative study of the karyotypes and electrophoretic patterns of Biomphalaria alexandrina and Bulinus truncatus and the ova of their corresponding trematode hosts. J Evol Biol Res 2011;3:22-28.
- 19. Abdel-Haleem AA. Comparative karyological studies on the two Egyptian schistosome vectors, Biomphalaria glabrata and B. alexandrina, with reference to chromosomal aberrations due to Za'ater plant. Internatl J Academic Sci Res 2013;1:1-7.
- 20. Natarajan R, Gismann A. Cytological studies of Planorbidae (Gastropoda: Basommatophora). I. Some African Planorbinae, Planorbininae and Bulininae. Malacologia 1965;2:239-251.
- 21. Burch JB. Chromosome of Pomatiopsis and Oncomelania. Am Malacol Un Ann Rep 1959, 1960;26:15.
Fig. 1Shell of Biomphalaria tenagophila from Rio de Janeiro, Brazil.

Fig. 2Metaphase chromosomes of Biomphalaria tenagophila (A) and karyotype constructed from A (B).
Table 1.Chromosome numbers and karyotypes of subfamily Biomphalarinae
Table 1.
|
Species |
Haploid no. |
Karyotypea
|
Source |
References |
|
Biomphalaria alexandria
|
18 |
|
Egypt |
Natarajan and Gismann [20] |
|
18 |
10M+4SM+2A+2T |
Egypt |
Abdel-Haleem [19] |
|
18 |
8M+8SM+2ST |
Egypt |
Bakry and Garhy [18] |
|
B. choanomphala
|
18 |
|
Tanzania |
Burch [16] |
|
B. glabrata
|
18 |
|
Puerto Rico |
Burch [21] |
|
18 |
10M+4SM+2A+2T |
USA |
Raghunathan [17] |
|
18 |
10M+4SM+2A+2T |
Egypt |
Abdel-Haleem [19] |
|
B. sudanica
|
18 |
|
Sudan |
Burch [21] |
|
B. tanganyicensis
|
18 |
|
Tanzania |
Natarajan and Gismann [20] |
|
B. pfeifferi
|
18 |
|
Tanzania South Africa |
Natarajan and Gismann [20] |
|
B. p. gaudi
|
18 |
|
Liberia, Ghana |
Natarajan and Gismann [20] |
|
B. p. madagascariensis
|
18 |
|
Madagascar |
Natarajan and Gismann [20] |
|
B. tenagophila
|
18 |
7M+8SM+3ST |
Brazil |
This study |
|
B. truncatus
|
18 |
10M+4SM+2ST+2T |
Egypt |
Bakry and Garhy [18] |
Table 2.Total lengths (μm) and relative lengths of chromosomes of Biomphalaria tenagophila
Table 2.
|
Chromosome |
Total length ± SE |
Relative length ± SE |
Type |
|
1 |
6.4 ± 0.21 |
9.3 ± 0.42 |
M |
|
2 |
6.2 ± 0.80 |
9.0 ± 0.35 |
M |
|
3 |
5.5 ± 0.45 |
8.0 ± 0.63 |
M |
|
4 |
5.3 ± 0.11 |
7.7 ± 0.16 |
ST |
|
5 |
4.8 ± 0.43 |
7.0 ± 0.65 |
SM |
|
6 |
4.6 ± 0.19 |
6.7 ± 0.32 |
M |
|
7 |
4.4 ± 0.10 |
6.4 ± 0.19 |
SM |
|
8 |
4.2 ± 0.14 |
6.1 ± 0.27 |
M |
|
9 |
4.0 ± 0.10 |
5.8 ± 0.54 |
SM |
|
10 |
3.9 ± 0.08 |
5.7 ± 0.35 |
M |
|
11 |
3.7 ± 0.23 |
5.4 ± 0.15 |
SM |
|
12 |
3.6 ± 0.07 |
5.2 ± 0.23 |
SM |
|
13 |
3.4 ± 0.13 |
4.9 ± 0.13 |
M |
|
14 |
3.2 ± 0.11 |
4.7 ± 0.16 |
SM |
|
15 |
3.0 ± 0.15 |
4.4 ± 0.33 |
SM |
|
16 |
2.8 ± 0.12 |
4.1 ± 0.12 |
ST |
|
17 |
2.6 ± 0.09 |
3.8 ± 0.08 |
SM |
|
18 |
2.4 ± 0.26 |
3.4 ± 0.21 |
ST |
Citations
Citations to this article as recorded by