Abstract
Variant surface antigens (VSAs) encoded by pir families are considered to be the key proteins used by many Plasmodium spp. to escape the host immune system by antigenic variation. This attribute of VSAs is a critical issue in the development of a novel vaccine. In this regard, a population genetic study of vir genes from Plasmodium vivax was performed in the Republic of Korea (ROK). Eighty-five venous blood samples and 4 of the vir genes, namely vir 27, vir 21, vir 12, and vir 4, were selected for study. The number of segregating sites (S), number of haplotypes (H), haplotype diversity (Hd), DNA diversity (π and Θw), and Tajima’s D test value were conducted. Phylogenetic trees of each gene were constructed. The vir 21 (S=143, H=22, Hd=0.827) was the most genetically diverse gene, and the vir 4 (S=6, H=4, Hd=0.556) was the opposite one. Tajima’s D values for vir 27 (1.08530, P>0.1), vir 12 (2.89007, P<0.01), and vir 21 (0.40782, P>0.1) were positive, and that of vir 4 (−1.32162, P>0.1) was negative. All phylogenetic trees showed 2 clades with no particular branching according to the geographical differences and cluster. This study is the first survey on the vir genes in ROK, providing information on the genetic level. The sample sequences from vir 4 showed a clear difference to the Sal-1 reference gene sequence, whereas they were very similar to those from Indian isolates.
-
Key words: Plasmodium vivax, vir gene, genetic diversity, the Republic of Korea
INTRODUCTION
Plasmodium vivax is the most prevalent human malaria parasite causing most of the cases occurring outside of sub-Saharan Africa [
1,
2]. Vivax malaria only infects host reticulocytes, the precursor of erythrocytes, whereas
Plasmodium falciparum infects all stages of erythrocytes [
3]. This narrow range of infection ability of
P. vivax gives it the stereotype of mild-type malaria. Until now, vivax malaria has been overlooked because of its mild characteristics compared with severe falciparum malaria [
4]. However, contrary to the benign reputation of vivax malaria, its clinical manifestations are not very mild, inciting chronic infection with hypoproteinemia, edema, and coma. Furthermore, the patients with vivax malaria and concurrent nutrient deficiency can suffer from splenic rupture, which and at last, may result in death [
5–
7].
Since 1993,
P. vivax has been consistently present in the northern areas of Gyunggido (Province) and Gangwon-do, Republic of Korea (ROK) [
8,
9]. Soldiers (30.5%) and veterans (25.1%) of the ROK make up over a half of the malaria patients in the country [
10]. Residents (including civilians and soldiers) living near the demilitarized zone (DMZ) on the northern border of the ROK have also been infected with
P. vivax. In this regard, chloroquine and primaquine have been used as a prophylactic measurement for the soldiers of the endemic area. However, such massive chemoprophylaxis using both drugs leads to a concern on an occurrence of antimalarial drug resistant
P. vivax in ROK.
Ever since chloroquine-resistant
P. vivax (CRPv) was first reported in Papua New Guinea [
11], such strain has been reported in many other places, including India, Vietnam, and even Bolivia of South America [
4,
12–
14], with spread occurring globally within decades. This expansion of CRPv demonstrates that once an antimalarial drug-resistant strain develops, it would soon become an international problem. Therefore, the development of malaria vaccines is receiving an attention as an alternative way to prevent vivax malaria.
By virtue of their antigenic variation, variant surface antigens (VSAs) of many
Plasmodium spp. are considered to be the key proteins used to escape from the host immune system through antigenic variations [
15]. These proteins are coded by multigene families located on telomeric and subtelomeric regions of the parasite’s chromosomes. The
var and variant interspersed repeats (
vir) gene superfamilies have been recognized in
P. falciparum and
P. vivax, respectively [
16]. The 346
vir genes are divided into 12 subfamilies, named A to L [
17,
18], and are reported to be related to host immune system evasion [
5]. In addition, the
vir proteins were revealed to be exported to the infected erythrocyte’s surface and partially induce the adherence of
P. vivax-infected reticulocytes to the endothelial cell receptor [
19,
20]. All together, these data suggest that the VSAs encoded by the
vir genes could be candidate targets for vaccine development, but the lack of an in vitro culture system for
P. vivax and the variant nature of VSAs have been an obstacle to the development of a potent vaccine [
21].
Trials for developing vaccines against
var-encoded proteins have been partially established, dismissing the skepticism over caused by the variance of
var proteins [
22]. By disrupting the erythrocyte rosettes, protection against sequestration could be established [
23]. Like the
var genes,
vir genes are potential targets for vaccination against vivax malaria [
17]. Although researching the genetic variation of VSAs is critical in the development of a novel vaccine, there has been no investigation on the subject until now in the ROK. Previously, Gupta et al. [
7,
24,
25] investigated the diversity of
P. vivax populations from various regions in India with
vir genes. Here, we performed a population genetic study in ROK. Among
vir genes, the
vir 27,
vir 21,
vir 12, and
vir 4 sequences on the subtelomeric area were more specifically analyzed. This study will provide a better understanding of the diversity of
vir genes.
MATERIALS AND METHODS
Ethics
The study was approved by the Ethics Committee of the Kyungpook National University. An approval form was used to obtain written informed consent from each participant. Each participant also consented to provide a 5-ml blood sample.
Sample collection
A total 85 venous blood samples were collected in the northern part of ROK, which are already known as malarial endemic regions [
9], from 2011 to 2013. The samples included 5 from Gimpo (GP), 2 from Paju (PJ), and 15 from Cheorwon (CW) in 2011; 6 from GP, 23 from PJ, and 7 from CW in 2012; and 10 from PJ and 17 from CW in 2013 (
Fig. 1). The collection criterion was the patient living in the study area who showed tertian fever. The blood samples were kept in EDTA-treated tube and cooled in −70°C.
Rapid diagnostic test kits (RDTs) (NanoSign Malaria P.f/P.v from Bioland, Cheonan, Korea, and SD Bioline Ag P.f/pan Rapid from SD, Hwaseong-si, Korea) were used for the first screening. Subsequently, microscopic examinations and nested PCR were performed to confirm the tentative diagnosis of vivax malaria. The microscopic tests were performed with a thick and thin blood films stained with Giemsa, and nested PCR targeting 18S rRNA was performed as previously described [
26].
Genomic DNA of P. vivax was extracted from 200 μl of each blood sample using a QIAamp DNA Mini Kit (Qiagen, Valencia, California, USA) following the manufacturer’s instructions. The extracted DNA was preserved in −70°C until required.
PCR amplification of vir genes
Four of the
vir genes, named
vir 27,
vir 21,
vir 12, and
vir 4, were selected (
Table 1). Compared with
vir 27, which belongs to subfamily I,
vir 4 of subfamily C and
vir 21 of subfamily B have variants with more polymorphic and distributed conserved blocks within the sequences, whereas
vir 12 of subfamily E is considered to be more conserved but shown high diversity among previous studies conducted on Indian isolates [
24,
25]. These
P. vivax sequences were amplified according to the following protocol. Each
vir gene was amplified by primary and nested PCR with 4 primers for
vir 27,
vir 21, and
vir 4, and 3 primers for
vir 12, respectively. The primary PCR mixture contained 2.5 μl of 10×buffer, 2 μl of dNTPs, 1 μl of forward primer, 1 μl of reverse primer, 0.25 μl of Ex taq polymerase (Takara, Shiga, Japan), 1 μl of DNA template, and distilled water to make up the volume to 25 μl. The semi-nested PCR was performed with a similar ratio of components but with 2 μl of DNA template. The primary PCR cycle was 95°C for 5 min; 35 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min; and a final extension of 72°C for 7 min. The semi-nested PCR had 30 cycles of the annealing step. Electrophoresis was performed on a 1.5% agarose gel with 50 mg/ml ethidium bromide. Ladders used were 100 bp for
vir 27,
vir 4, and
vir 21, and 1 kb for
vir 12.
Amplicons were purified with an AccuPrep® Gel Purification Kit (Bioneer, Daejeon, Korea) for cloning, according to the manufacturer’s instructions. After gel purification, the PCR products were ligated into the pGEM-T TA cloning vector (Promega, Madison, Wisconsin, USA) and transformed into Escherichia coli DH5α competent cells. Five colonies with the recombinant plasmids per each vir gene were selected, and then, sequenced with the pUC/M13 forward and reverse primers by using a Big-Dye Terminator ver. 3.1 sequencing kit (Applied Biosystems, Foster City, California, USA) with an automatic ABI3730 DNA Analyzer sequencer (Applied Biosystems).
Sequence analysis
Acquired sequences were aligned by the Clustal Omega multiple alignment function of BioEdit [
27]. The segregating sites (S), the number of haplotypes (H), haplotype diversity (Hd), the average number of pairwise nucleotide differences (π), and the total number of segregating sites (Θ
w) were evaluated by using DnaSP ver. 5.10.01 [
28].
The π and the Θ
w values were used to estimate the genetic diversity. The π value indicates the randomness of the sample, whereas the Θ
w value indicates the equilibrium of gene segregating sites in a population [
29,
30]. These 2 measures are also compared as a test of neutrality. Tajima’s D test of neutrality was done with the sliding window option (window length, 100 bp; step size, 25 bp) for distinguishing whether the
P. vivax DNA evolved randomly, neutrally, or not, including balancing selection or directional selection [
31]. This test was calculated with DnaSP ver. 5.10.01. Any differences in the mean values were considered to be statistically significant when at
P<0.05.
The
vir genes of
P. vivax Sal-1 (GenBank accession nos. AAKM01000041.2 for
vir 27, AAKM01000104.1 for
vir 4, AAKM01000016.1 for
vir 12, and AAKM01000003.1 for
vir 21) were used as the reference. Using the minimum-evolution method and maximum composite likelihood model option, the ROK
vir gene sequences were compared with the Sal-1 references, and phylogenetic trees were constructed using MEGA 7.0 [
32]. Each node in the trees was supported by bootstrap analysis with 1,000 repetitions. The subjects of the phylogenetic trees are labeled with the name of the gene, sample number, collected year, and region.
RESULTS
Nested-PCR results
The nested-PCR results were evaluated according to the year and region collected as shown in
Table 2. The
vir 27 gene revealed the highest PCR-positive rate (72.9%), followed by
vir 12 (65.7%),
vir 21 (46.7%), and
vir 4 (31.3%).
Specifically, vir 27 was amplified in 59.1% (13/22), 80.6% (29/36), and 74.1% (20/27) of the samples collected in 2011, 2012, and 2013, respectively. According to the sample collection year, samples from 2012 showed the highest PCR-positive rate (80.6%). Per classification by region, the GP samples had the highest rate of PCR-positive results (81.8%).
For vir 21, 43 samples that were composed of 50.0% (11/22), 47.2% (17/36), and 55.6% (15/27) of the samples collected in 2011, 2012, and 2013, respectively, were amplified indicating that these samples were PCR positive. Gene was most prevalent in the samples of 2011 and 2013. Samples from PJ and CW had the highest PCR-positive rates (54.3% and 51.3%, respectively).
For vir 12, 50.0% (11/22), 83.3% (30/36), and 66.7% (18/27) of the samples collected in 2011, 2012, and 2013, respectively, were PCR positive, showing 2012 to have the highest prevalence for this gene. Samples from PJ showed the highest PCR-positive rate at 74.3%.
For vir 4, 22.7% (5/22), 36.1% (13/36), and 29.6% (8/27) of the samples collected in 2011, 2012, and 2013, respectively, were PCR positive, again with 2012 having the highest rate for this gene. The PCR-positive rate of the GP samples was higher than that of PJ and CW (36.4%, 25.7%, and 33.3%, respectively). Gene vir 4 showed the lowest positive rate on PCR.
Variable surface protein 27 gene
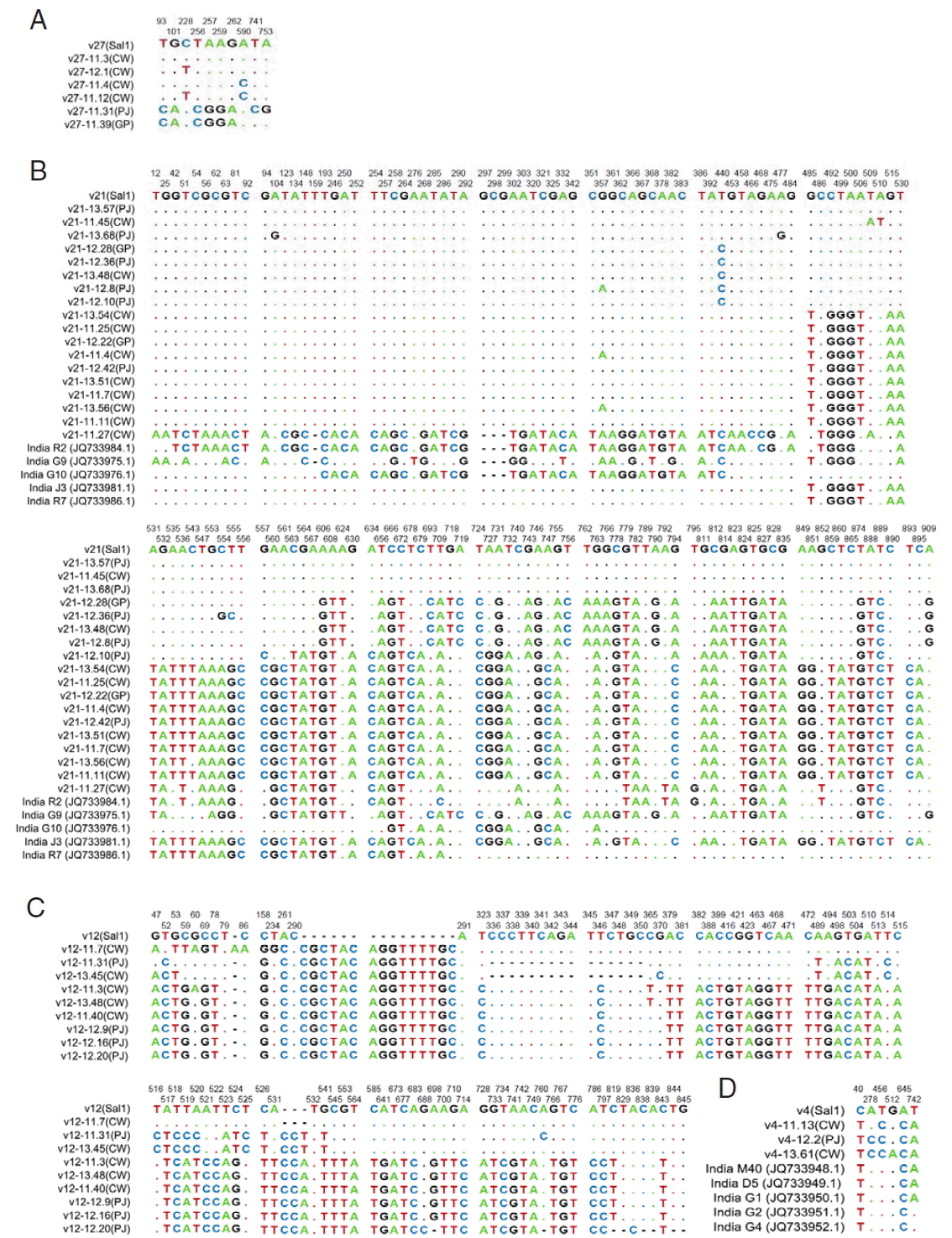
Compared with the Sal-1 reference sequence, 62
vir 27 genes showed 10 S, 2 of them being singleton variable sites and 8 of them being parsimony-informative sites. The nucleotide variations T93C, G101A, C228T, T256C, A257G, A259G, G262A, A590C, T741C, and A753G were separately found (
Fig. 2A). The sequences of
vir 27 genes are available in GenBank (nos. KY608472-608521). The H value for
vir 27 was 6, and the Hd value was 0.774.
The
vir 27 gene from the 62 ROK samples was divided into 2 distinct clades in phylogenetic analysis. One clade was comprised of 45 samples, including Sal-1 reference gene, and was divided into several subclades which consisted of 18 samples. In the clade of 45 samples, the main subclade was composed of 21 samples, and the other subclade was further composed of 2 clades with 10 samples each and 1 clade with 4 samples (
Supplement Fig. 3A).
There were 43 samples PCR positive for
vir 21 genes. A total of 143 S were found, including 55 singleton sites with 2 variants, only 1 singleton sites with 3 variants, 84 parsimony-informative sites with 2 variants, and 3 parsimony-informative sites with 3 variants (
Fig. 2B). The sequences of
vir 21 genes are available in GenBank (nos. KY608420-608471). The H and Hd values were 22 and 0.827, respectively. Nucleotide diversity values were high for
vir 21 (π=0.04165, Θ
w=0.03746) compared to those of other
vir genes (
Table 3;
Supplement Fig. 1). Tajima’s D values for
vir 21 were positive (0.40782,
P>0.1;
Tables 3;
Supplement Fig. 2).
In the phylogenetic analysis of
vir 21, an individual sample collected from CW (v21-11.27) was found. This sample was subjected to BLAST analysis whereupon it showed 99% (952/961) identity with
P. vivax clone R2_21 (GenBank no. JQ733984.1) from an Indian isolate [
24]. Otherwise, the difference between v21-11.27 and the Sal-1 reference gene
vir 21 was 92% (882/961). With 5 Indian samples R2, R7, G9, G10, and J3 (GeneBank nos. JQ733984.1, JQ733986.1, JQ733975.1, JQ733976.1, and JQ733981.1, respectively), 2 clades were revealed on phylogenetic tree with almost the same sample sizes of 19 and 20, respectively. Compared with the clade of 19 samples, 20-sample clade had several more subclades (
Supplement Fig. 3B).
Among the 59 samples PCR positive for
vir 12, there were 74 S. Six were singleton sites with 2 variants, and 68 were parsimony-informative with 2 variants, and 4 were parsimony-informative sites with 3 variants. Distinct nucleotide insertion was found at site 291, CGCTACAGGTTTTGC, and a number of nucleotides changes were also found (
Fig. 2C). The sequences of
vir 12 genes are available in GenBank (nos. KY608364-608419). The values of H and Hd were 9 and 0.795, respectively. Nucleotide diversity values (π and Θ
w) of
vir 12 were 0.03640 and 0.01979, respectively. Tajima’s D value for
vir 12 was 2.89007 (
P<0.01;
Table 3;
Supplement Fig. 2).
The phylogenetic tree of
vir 12 consisted of 2 separate clades. The main branch contained 3 sub-branches of 2, 15, and 17 samples, respectively. The Sal-1 reference strain was located in a second branch of 25 samples, which was divided into 2 sub-branches: 1 with 12 samples (separated with Sal-1) and the other with 12 samples (
Supplement Fig. 3C).
In the case of the 26
vir 4 gene sequences studied, 6 were S. Among nucleotide sequences, changes were found (A278C in 10 isolates, G512A in 1 isolate, and C40T, T456C, A645C, and T742A in all the isolates, respectively) (
Fig. 2D). The sequences of
vir 4 genes are available in GenBank (nos. KY608341-608363). The H and Hd values were 4 and 0.556, respectively. In contrast to other
vir genes, the π values of
vir 4 were lower than the Θ
w values (π=0.00064, Θ
w=0.00116). Moreover,
vir 4 had the only negative Tajima’s D value region (
Table 3;
Supplement Fig. 2).
There was a distinct difference between the
vir 4 genes of the 26 ROK samples and that of the Sal-1 reference strain. The ROK clade had 2 groups consisting of 16 samples and 10 samples, respectively. BLAST alignment of the ROK
vir 4 genes resulted in 93–97% identity with
P. vivax clone M40_4 (GenBank no.: JQ733948.1), D5 (JQ733949.1), G1 (JQ7339550.1), G2 (JQ733951.1), and G4 (JQ733952.1) from Indian isolate. With the sequences of Indian isolates, there were common mutation with Korean isolates in T742A with M40, D5, and G1. However, D5 isolate have the only mutation in G871A [
24]. Otherwise, the differences between the ROK and Sal-1
vir 4 genes were 73–79% (
Fig. 3).
The π and Θ
w values were calculated separately for each
vir gene and region (
Table 4). The average π and Θ
w values of all
vir genes were 0.02027 and 0.01503, respectively. The π values were generally higher than the Θ
w values except for
vir 4 (π= 0.00064, Θ
w=0.00116). In the regional analysis, similar to the average values, the π values were higher than the Θ
w values in PJ and CW, but reverse was true for the GP samples.
DISCUSSION
Plasmodium spp. have several mechanisms for host immune evasion [
33]. One of them is the
pir multigene superfmily, such as
kir, bir, cir, and
yir families encoded by
Plasmodium knowlesi,
Plasmodium berghei, Plasmodium chaubadi, and
Plasmodium yoelii [
24,
34–
36]. Especially in
P. falciparum, these genes already studied that encoded several functional proteins known as
stevor/rif and
var in
P. falciparum [
24]. This attribute of avoiding host’s immune system can affect the severity of malaria infection in falciparum malaria by delay in immune acquisition, cytoadherence, and rosetting phenomenon [
37]. A recent study showed that several
vir genes have some relation with pathogenesis of vivax malaria [
7]. Furthermore,
cir of
P. chaubadi have functional role in blood stage malaria [
35].
In the present study, analysis of the
vir 27 sequences of all the samples revealed that this gene is more conserved than
vir 21 and
vir 12, having the same nucleotide sequences as
vir 27 of Sal-1 except for the mutations of 10 nucleotides. Gene
vir 21 was the most polymorphic, as proven in the previous research [
17]. In an earlier research,
vir 12 was described as being the most conserved gene [
17]. However, in our ROK samples,
vir 12 was the second most polymorphic gene with many segregating sites, and high haplotype and nucleotide diversity values. All sequences of
vir 4 (a member of
vir subfamily C that is considered to be the most diverse) differed from
vir 4 of Sal-1 at 6 mutation points. In spite of the reputation of
vir 4 as the more polymorphic gene than others, this sequences from ROK seemed to be well conserved. It might be already mutated before introduction in the ROK.
The number of segregating sites ranged from as low as 6 (
vir 4) to as high as 143 (
vir 21). The segregating sites induced variant changes into amino acid sequences of the samples. Such changes in amino acid sequences might have affected the antigenicity of
P. vivax, evading the immune system of the host. However, it is not so clear that how
vir gene superfamilies’ really work on the protozoal dodge against the host attack and result in the clinical manifestation of vivax malaria [
7].
Diversity parameters were similar to the number of segregating sites. First, the average number of H of all 4
vir genes was 10.25, and the average Hd value was 0.738. Gene
vir 21 (H=22, Hd=0.827) was the most genetically diverse, followed by
vir 12 (H=9, Hd=0.795),
vir 27 (H=6, Hd=0.774), and
vir 4 (H=6, Hd=0.556,). The H and Hd values of
vir 4 were comparatively lower than the others, alike to a previous study in India [
25]. Second, the π and Θ
w values of
vir genes were variable from high to low. It might indicate that there was ongoing molecular mutation among
vir genes of the ROK malaria isolates.
Tajima’s D test was computed for analyzing the natural selection occurrences of the
P. vivax vir genes from the ROK samples. A positive Tajima D value indicates that an increase of polymorphism, a decrease of population size, and/or balancing selection can occur. A negative D value means a low frequency of polymorphism, an expansion of population size, and/or purifying selection. The
vir 4 gene (−1.32162,
P>0.1) had the only negative value, indicating that this gene might be under unfavorable circumstance for survival and/or in the process of being purged from the genetic pool via population expansion, an effect that increases with a lower mutation rate but decreases with a higher recombination rate [
38]. With this result and the
vir 4 gene’s low PCR-positive rate, the
vir 4 may be disprefered to
P. vivax existing in the endemic region of ROK. Tajima’s D values for
vir 27 (1.08530,
P>0.1),
vir 12 (2.89007,
P<0.01), and
vir 21 (0.40782,
P>0.1) were all positive, meaning a population contraction of these genes and a process of balancing selection [
39]. Moreover, Tajima’s D value for
vir 12 was statistically significant, indicating that among 4
vir genes this gene is under balancing selection and/or decreasing population size. In addition, such value means that
vir 12 may be the major target of the host immune system in ROK. In the regional analysis, only
vir 27 samples from GP had negative Tajima’s D values differ from the Tajima’s D test result of whole samples (−0.92403). In this case,
vir 27 samples from GP were considered as under purifying selection, or expanding population size situation. However, it is hard to make a conclusion, because there were only 11 case collected. So, further studies will be needed. With the result of the
vir 21 and
vir 4 genes, there were some differences between few ROK isolates and Sal-1
vir genes. Also, those isolates were very similar to the
vir genes of Indian isolates. This inferred that
P. vivax malaria in the ROK might have some relationship with
P. vivax in India. However, it is hard to clarifying this relationship due to lack of knowledge about
vir gene superfamily.
In the current study, all phylogenetic trees showed clear 2 clades same as previous studies [
24,
25]. However, because of the randomly scattered sample distribution, there was no particular branching according to the geographical differences and hard to find obvious cluster.
In conclusion, this study was the first survey about the vir genes in the ROK, providing information on the genetic level. Among the 4 vir genes studied, the vir 27 was relatively conserved and expressed widely. Next, the most divergent was vir 12 whereas the most conserved was vir 4. However, the ROK sample sequences of some vir 21 and vir 4 showed a clear difference with the Sal-1 reference gene sequence, but were very similar to the vir 21 and vir 4 genes of Indian isolates. Moreover, several changes on the amino acid level were revealed. Nevertheless, it was hard to find some trend among the samples according to the period, because the study was conducted only for 3 years. Also, the revelation of the specific function of vir gene is not yet sufficient; therefore, applying VSAs from vir genes in practical areas is only at the beginning stages, and VSA variances could be an obstacle to achieving a certain product. With these results, however, the information boundary surrounding vir genes in the ROK was expanded, thereby advancing the first steps of the buildup the information basis. So, this kind of study about vir genes is needed in order to understand vivax malaria, the largest parasitic foe of healthcare.
Notes
-
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supplementary materials
Supplemental Fig. 1.
Sliding window plot of nucleotide diversity per site (π) at vir genes (vir 27, vir 21, vir 12, and vir 4). The π values were calculated on DnaSP with window length 100 bp and step size of 25 bp.
Supplemental Fig. 2.
Sliding window plot of Tajima’s D using a window size of 100 nucleotides and including each 62, 43, 59, and 26 alleles of vir 27, vir 21, vir 12, and vir 4. Positive values generally indicate that balancing selection is acting to maintain alleles at intermediate frequencies.
Supplemental Fig. 3.
The phylogenetic trees were conducted by minimum evolution method. The sum of branch lengths for each optimal tree were (A) vir 27=0.01169659. The phylogenetic trees were conducted by minimum evolution method. The sum of branch lengths for each optimal tree were (B) vir 21=0.17133662. The phylogenetic trees were conducted by Minimum Evolution method. The sum of branch lengths for each optimal tree were (C) vir 12=0.09495926.
ACKNOWLEDGMENTS
This work was supported by Biomedical Research Institute grant, Kyungpook National University Hospital (2014). We are grateful to all blood donors and the staff of the Public Health Centers and hospitals in the ROK.
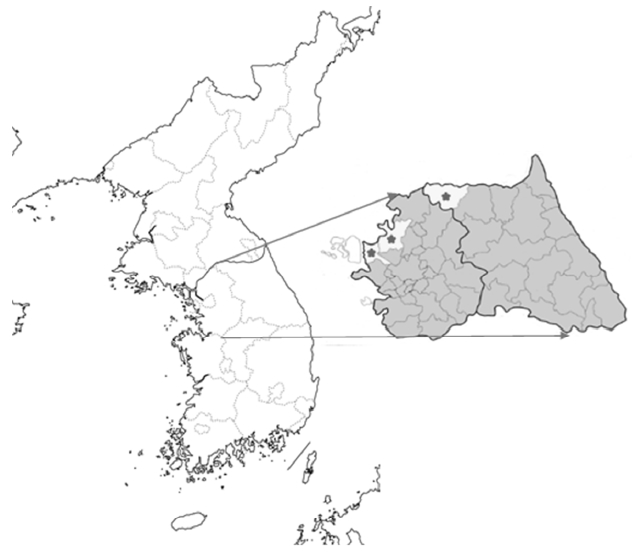
Fig. 1Blood collection regions. From left to right, Gimpo (GP), Paju (PJ), and Choerwon (CW) are denoted by asterisks.
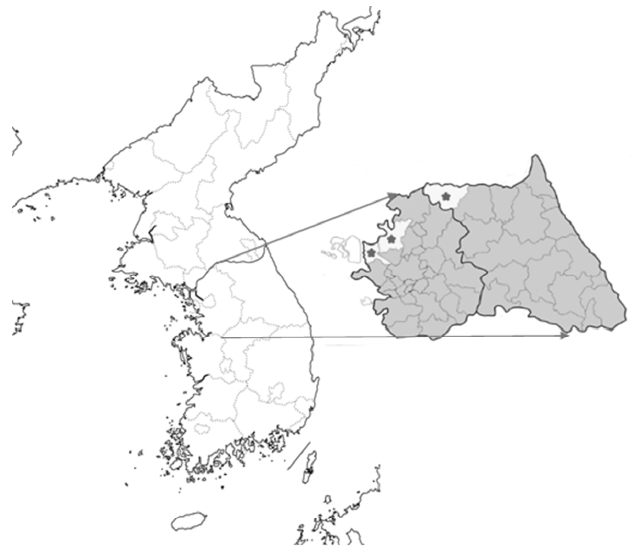
Fig. 2Sequence polymorphism of vir genes in the ROK P. vivax isolates. The polymorphic nucleotides were compared to that of Sal-1 (GenBank no.: AAKM01000041.2). (A) Sequences of vir 27 gene of isolates. (B) Sequences of vir 21 gene of ROK isolates compared with 5 Indian isolates. (C) Sequences of vir 12 gene of isolates. (D) Sequences of vir 4 gene of ROK and Indian isolates. The listed sequences are indicated by dots for identical nucleotides with sequences of Sal-1, and hyphens for gaps. The locations of polymorphic sites are represented above and the nucleotide changes are marked with colors.

Fig. 3The phylogenetic tree of vir 4 was inferred using the minimum evolution method. The optimal tree with the sum of branch length=0.00535425 is shown.

Table 1Information of PCR ingredients used in this study
Table 1
|
PCR primers |
Primer sequence |
Annealing temperature (°C) |
Subfamilya
|
Gene lengthb (bp) |
Accession no. |
|
vir 27 gene: PVX 102630 |
55 |
I |
1,255 |
AAKM01000041.2 |
|
v27_1 |
TGGCATTACACTTAGCGGTAT |
|
|
|
|
|
v27_4 |
TGGTGTATCTGTGTGAAGATTTG |
|
|
|
|
|
v27_2 |
CATTTTGAGCACGTTCTCCTT |
|
|
|
|
|
v27_3 |
TCAAGTTGTAAGGTGATCAATGAAA |
|
|
|
|
|
|
vir 21 gene: PVX 115480 |
|
B |
974 |
AAKM01000003.1 |
|
v21_1 |
ACAACTCATTGGTATTTACATTTATGA |
55 |
|
|
|
|
v21_4 |
TGCGAAAAATAAGACGCAAA |
|
|
|
|
|
v21_2 |
TTCAAAGCATAAAATTCGCACT |
|
|
|
|
|
v21_3 |
TCCCAAAATAAAGGCAAGGTT |
|
|
|
|
|
|
vir 12 gene: PVX 097525 |
|
E |
2,548 |
AAKM01000016.1 |
|
v12_1 |
AAATATTCAAACAATGGCAATACA |
55 |
|
|
|
|
v12_3 |
ACTTCCCGTGCAGGTATTTC |
|
|
|
|
|
v12_2 |
TGACATAGGTAACGCATAATATAGCA |
|
|
|
|
|
|
vir 4 gene: PVX 021680 |
|
C |
1,314 |
AAKM01000104.1 |
|
v4_1 |
CGAAATATACTTGTCTTAACTGGTTG |
55 |
|
|
|
|
v4_4 |
CAGAATGGCATCTGTTTATGC |
|
|
|
|
|
v4_2 |
GACCAATATTTTGGGCAGGTT |
|
|
|
|
|
v4_3 |
AGGTAGAATTGCGGCTCAGA |
|
|
|
|
Table 2PCR results of venous blood samples collected from malaria patients according to year of collection and region
Table 2
|
Year |
Region |
Vir 27 |
Vir 21 |
Vir 12 |
Vir 4 |
|
|
|
|
|
+ (%) |
− (%) |
+ (%) |
− (%) |
+ (%) |
− (%) |
+ (%) |
− (%) |
|
2011 |
Gimpo (GP) |
5 (22.7) |
0 (0.0) |
1 (20.0) |
4 (80.0) |
2 (40.0) |
3 (60.0) |
2 (40.0) |
3 (60.0) |
|
Paju (PJ) |
1 (4.6) |
1 (4.6) |
1 (50.0) |
1 (50.0) |
1 (50.0) |
1 (50.0) |
0 (0.0) |
2 (100.0) |
|
Cheorwon (CW) |
7 (31.8) |
8 (36.3) |
9 (60.0) |
6 (40.0) |
8 (53.3) |
7 (46.7) |
3 (20.0) |
12 (80.0) |
|
Total |
13 (59.1) |
9 (40.9) |
11 (50.0) |
11 (50.0) |
11 (50.0) |
11 (50.0) |
5 (22.7) |
17 (77.3) |
|
|
2012 |
Gimpo |
4 (66.6) |
2 (33.3) |
3 (50.0) |
3 (50.0) |
5 (83.3) |
1 (16.7) |
2 (33.3) |
4 (66.7) |
|
Paju |
18 (78.3) |
5 (21,7) |
12 (52.2) |
11 (47.8) |
19 (82.6) |
4 (17.4) |
6 (26.1) |
17 (73.9) |
|
Cheorwon |
7 (100.0) |
0 (0.0) |
2 (28.6) |
5 (71.4) |
6 (85.7) |
1 (14.3) |
5 (71.4) |
2 (28.6) |
|
Total |
29 (80.6) |
7 (20.4) |
17 (47.2) |
19 (52.8) |
30 (83.3) |
6 (16.7) |
13 (36.1) |
23 (63.9) |
|
|
2013 |
Gimpo |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Paju |
6 (60.0) |
4 (40.0) |
6 (60.0) |
4 (40.0) |
6 (60.0) |
4 (40.0) |
3 (30.0) |
7 (70.0) |
|
Cheorwon |
14 (82.4) |
3 (17.6) |
9 (52.9) |
8 (47.1) |
12(70.6) |
5 (29.4) |
5 (29.4) |
12 (70.6) |
|
Total |
20 (74.1) |
7 (25.9) |
15 (55.6) |
12 (44.4) |
18 (66.7) |
9 (33.3) |
8 (29.6) |
19 (70.4) |
|
|
Total |
|
62 (72.9) |
23 (27.1) |
43 (46.7) |
42 (53.3) |
59 (65.7) |
26 (34.3) |
26 (31.3) |
59 (68.7) |
Table 3Summary statistics for the 4 vir genes in gene level
Table 3
|
Vir genes (No. of analyzed genes) |
Sequence length |
Segregating sites (S) |
No. of haplotypes (H) |
Haplotype diversity (Hd) |
Nucleotide diversity |
Tajima’s D test |
|
π |
Θw
|
|
Vir 27 (62) |
1,255 bp |
10 |
6 |
0.774 |
0.00238 |
0.00171 |
1.08530 |
|
Vir 21 (43) |
974 bp |
143 |
22 |
0.827 |
0.04165 |
0.03746 |
0.40782 |
|
Vir 12 (59) |
1,323 bp |
74 |
9 |
0.795 |
0.03640 |
0.01979 |
2.89007a
|
|
Vir 4 (26) |
1,314 bp |
6 |
4 |
0.556 |
0.00064 |
0.00116 |
−1.32162 |
|
Average |
|
58.25 |
10.25 |
0.738 |
0.02027 |
0.01503 |
0.76539 |
Table 4Summary statistics for 4 vir genes with population
Table 4
|
Region |
Vir genes (No. of analyzed genes) |
Sequence length |
Segregating sites (S) |
No. of haplotypes (H) |
Haplotype diversity (Hd) |
Nucleotide diversity |
Tajima’s D test |
|
|
π |
Θw
|
|
Gimpo (GP) |
Vir 27 (9) |
1,255 bp |
8 |
4 |
0.778 |
0.00178 |
0.00227 |
−0.92403 |
|
Vir 21 (4) |
974 bp |
83 |
5 |
1.000 |
0.04544 |
0.04426 |
0.20329 |
|
Vir 12 (7) |
1,323 bp |
70 |
4 |
0.821 |
0.04043 |
0.03164 |
2.89007a
|
|
Vir 4 (4) |
1,314 bp |
4 |
2 |
0.400 |
0.00119 |
0.00143 |
−1.09380 |
|
|
Paju (PJ) |
Vir 27 (25) |
1,255 bp |
10 |
6 |
0.788 |
0.00293 |
0.00231 |
0.89090 |
|
Vir 21 (19) |
974 bp |
86 |
10 |
0.758 |
0.03489 |
0.02763 |
1.07769 |
|
Vir 12 (26) |
1,323 bp |
79 |
8 |
0.849 |
0.03658 |
0.02469 |
1.86160a
|
|
Vir 4 (9) |
1,314 bp |
5 |
3 |
0.644 |
0.00100 |
0.00132 |
−0.98485 |
|
|
Cheorwon (CW) |
Vir 27 (28) |
1,255 bp |
7 |
5 |
0.771 |
0.00236 |
0.00143 |
1.92777 |
|
Vir 21 (20) |
974 bp |
144 |
15 |
0.957 |
0.04800 |
0.04484 |
0.28890 |
|
Vir 12 (26) |
1,323 bp |
76 |
8 |
0.803 |
0.03766 |
0.02359 |
2.30545b
|
|
Vir 4 (13) |
1,314 bp |
6 |
4 |
0.659 |
0.00093 |
0.00141 |
−1.22712 |
References
- 1. Dalrymple U, Mappin B, Gething PW. Malaria mapping: understanding the global endemicity of falciparum and vivax malaria. BMC Med 2015;13:140.
- 2. World Health Organization. World malaria report 2015. Geneva, Switzerland.
- 3. Popovici J, Ménard D. Challenges in antimalarial drug treatment for vivax malaria control. Trends Mol Med 2015;21:776-788.
- 4. Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg 2007;77:79-87.
- 5. del Portillo HA, Lanzer M, Rodriguez-Malaga S, Zavala F, Fernandez-Becerra C. Variant genes and the spleen in Plasmodium vivax malaria. Int J Parasitol 2004;34:1547-1554.
- 6. World Health Organization. World malaria report 2013. Geneva, Switzerland. World Health Organization; 2013.
- 7. Gupta P, Sharma R, Chandra J, Kumar V, Singh R, Pande V, Singh V. Clinical manifestations and molecular mechanisms in the changing paradigm of vivax malaria in India. Infect Genet Evol 2016;39:317-324.
- 8. Kim HC, Pacha LA, Lee WJ, Lee JK, Gaydos JC, Sames WJ, Lee HC, Bradley K, Jeung GG, Tobler SK, Klein TA. Malaria in the Republic of Korea, 1993–2007. Variables related to re-emergence and persistence of Plasmodium vivax among Korean populations and U.S. forces in Korea. Mil Med 2009;174:762-769.
- 9. Kim JY, Goo YK, Zo YG, Ji SY, Trimarsanto H, To S, Clark TG, Price RN, Auburn S. Further evidence of increasing diversity of Plasmodium vivax in the Republic of Korea in recent years. PLoS One 2016;11:e0151514.
- 10. Linthicum KJ, Anyamba A, Killenbeck B, Lee WJ, Lee HC, Klein TA, Kim HC, Pavlin JA, Britch SC, Small J, Tucker CJ, Gaydos JC. Association of temperature and historical dynamics of malaria in the Republic of Korea, including reemergence in 1993. Mil Med 2014;179:806-814.
- 11. Rieckmann K, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine? Lancet 1989;2:1183-1184.
- 12. Dua VK, Kar PK, Sharma VP. Chloroquine resistant Plasmodium vivax malaria in India. Trop Med Int Health 1996;1:816-819.
- 13. Añez A, Moscoso M, Laguna Á, Garnica C, Melgar V, Cuba M, Gutierrez S, Ascaso C. Resistance of infection by Plasmodium vivax to chloroquine in Bolivia. Malaria J 2015;14:261.
- 14. Thanh PV, Hong NV, Van NV, Louisa M, Baird K, Xa NX, Peeters Grietens K, Hung le X, Duong TT, Rosanas-Urgell A, Speybroeck N, D’Alessandro U, Erhart A. Confirmed Plasmodium vivax resistance to chloroquine in central Vietnam. Antimicrob Agents Chemother 2015;59:7411-7419.
- 15. Fernandez-Becerra C, Pein O, de Oliveira TR, Yamamoto MM, Cassola AC, Rocha C, Soares IS, de Bragança Pereira CA, del Portillo HA. Variant proteins of Plasmodium vivax are not clonally expressed in natural infections. Mol Microbiol 2005;58:648-658.
- 16. Pasternak ND, Dzikowski R. PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum. Int J Biochem Cell Biol 2009;41:1463-1466.
- 17. Fernandez-Becerra C, Yamamoto MM, Vêncio RZ, Lacerda M, Rosanas-Urgell A, del Portillo HA. Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol 2009;25:44-51.
- 18. Lopez FJ, Bernabeu M, Fernandez-Becerra C, del Portillo HA. A new computational approach redefines the subtelomeric vir superfamily of Plasmodium vivax. BMC Genomics 2013;14:8.
- 19. Carvalho BO, Lopes SC, Nogueira PA, Orlandi PP, Bargieri DY, Blanco YC, Mamoni R, Leite JA, Rodrigues MM, Soares IS, Oliveira TR, Wunderlich G, Lacerda MV, del Portillo HA, Araújo MO, Russell B, Suwanarusk R, Snounou G, Rénia L, Costa FT. On the cytoadhesion of Plasmodium vivax-infected erythrocytes. J Infect Dis 2010;202:638-647.
- 20. Bernabeu M, Lopez FJ, Ferrer M, Martin-Jaular L, Razaname A, Corradin G, Maier AG, del Portillo HA, Fernandez-Becerra C. Functional analysis of Plasmodium vivax VIR proteins reveals different subcellular localizations and cytoadherence to the ICAM-1 endothelial receptor. Cell Microbiol 2012;14:386-400.
- 21. Arévalo-Herrera M, Chitnis C, Herrera S. Current status of Plasmodium vivax vaccine. Hum vaccin 2010;6:124-132.
- 22. Makobongo MO, Keegan B, Long CA, Miller LH. Immunization of Aotus monkeys with recombinant cysteine-rich interdomain region 1 alpha protects against severe disease during Plasmodium falciparum reinfection. J Infect Dis 2006;193:731-740.
- 23. Chen Q, Pettersson F, Vogt AM, Schmidt B, Ahuja S, Liljeström P, Wahlgren M. Immunization with PfEMP1-DBL1α generates antibodies that disrupt rosettes and protect against the sequestration of Plasmodium falciparum-infected erythrocytes. Vaccine 2004;22:2701-2712.
- 24. Gupta P, Das A, Singh OP, Ghosh SK, Singh V. Assessing the genetic diversity of the vir genes in Indian Plasmodium vivax population. Acta Trop 2012;124:133-139.
- 25. Gupta P, Pande V, Das A, Singh V. Genetic polymorphisms in VIR genes among Indian Plasmodium vivax populations. Korean J Parasitol 2014;52:557-564.
- 26. Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol 1993;61:315-320.
- 27. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95-98.
- 28. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009;25:1451-1452.
- 29. Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol 1975;7:256-276.
- 30. Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics 1983;105:437-460.
- 31. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989;123:585-595.
- 32. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-1874.
- 33. Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis 2009;9:555-566.
- 34. Janssen CS, Phillips RS, Turner CM, Barrett MP. Plasmodium interspersed repeats: the major multigene superfamily of malaria parasites. Nucleic Acids Res 2004;32:5712-5720.
- 35. Yam XY, Brugat T, Siau A, Lawton J, Wong DS, Farah A, Twang JS, Gao X, Langhorne J, Preiser PR. Characterization of the Plasmodium interspersed repeats (PIR) proteins of Plasmodium chabaudi indicates functional diversity. Sci Rep 2016;6:23449.
- 36. Jackson AP. Gene family phylogeny and the evolution of parasite cell surfaces. Mol Biochem Parasitol 2016;209:64-75.
- 37. A-Elgadir TM, Theander TG, Elghazali G, Nielsen MA, A-Elbasit IE, Adam I, Troye-Blomberg M, Elbashir MI, Giha HA. Determinants of variant surface antigen antibody response in severe Plasmodium falciparum malaria in an area of low and unstable malaria transmission. Scand J Immunol 2016;63:232-240.
- 38. Hudson RR, Kaplan NL. Deleterious background selection with recombination. Genetics 1995;141:1605-1617.
- 39. Kang JM, Ju HL, Kang YM, Lee DH, Moon SU, Sohn WM, Park JW, Kim TS, Na BK. Genetic polymorphism and natural selection in the C-terminal 42 kDa region of merozoite surface protein-1 among Plasmodium vivax Korean isolates. Malar J 2012;11:206.