Abstract
Three species of the families Viviparidae and Pleuroceridae, the first intermediate host of paragonimiasis, metagonimiasis and echinostomiasis were studied cytologically. The observed diploid chromosome number was as follows: Semisulcospira libertina 36, S. dolichostoma 34, and Viviparus rivularis 64. The mitotic chromosome complement of S. libertina has nine metacentric pairs and nine submetacentric pairs, and S. dolichostoma has three metacentric pairs and 14 submetacentric pairs of chromosomes. Viviparus rivularis showed two metacentric pairs and 30 submetacentric pairs of chromosomes.
-
Key words: chromosome numbers, karyotypes, Chinese mesogastropods, Viviparidae, Pleuroceridae
INTRODUCTION
Taxonomy of viviparid and pleurocerid snails which serve as intermediate hosts for several kinds of trematodes has been in a chaotic state for many years (
Burch, 1975). The systematics of the lower taxonomic categories still remains an enigma, both from the practical viewpoint of identifying specimens and in understanding the mechanisms of speciation and evolution (
Burch, 1968). In mollusca, the literatures on karyotype analysis are not abundant due to difficulties of obtaining mitotic fields with enough quality to carry out chromosome studies. "Melaniid" (=Pleurocerid) snails are among the most common and widely dis-tributed groups of freshwater mollusks in the world, however studies concerning their chromosomes are rare. Until now, the chromosomes of six species have been described among the poorly understood genera
Semisulcospira and
Viviparus occurring in China.
The purpose of this study was to detect the chromosome numbers and to identify the karyotypes of two pleurocerid and one viviparid Chinese snails, which serve as molluscan intermediate hosts for paragonimiasis, metagonimiasis and echinostomiasis.
MATERIALS AND METHODS
The snails in this study were collected from Houwei and Suncun Villages, Jinde County, Anhui Province, China, in October 1997. Chromosome preparations were made from gonadal tissues by the air-drying method (
Park, 1994). The materials examined were gonads in active stages of gametogenesis from six specimens of
Semisulcospira libertina, five specimens of
S. dolichostoma and three specimens of
Viviparus rivularis. To observe morphological features of the chromosomes, karyotypes, relative and total lengths of the mitotic metaphase chromosomes were measured. Nomenclature of morphological types of chromosome was adopted from Levan et al. (
1964).
RESULTS
Pleuroceridae
Semisulcospira libertina
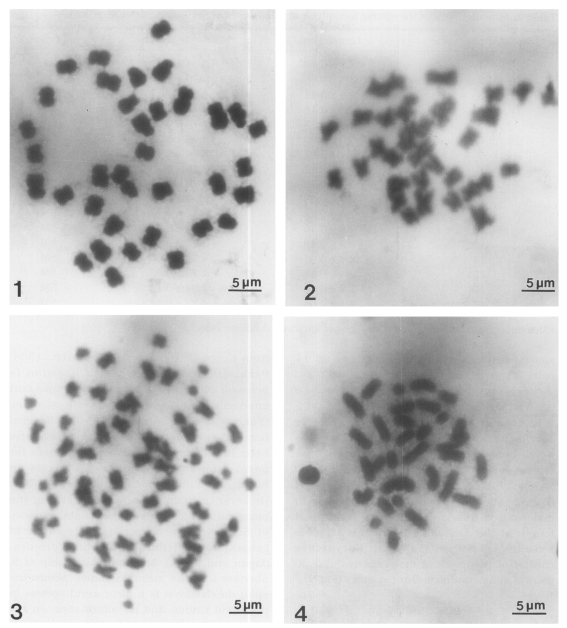
Chromosomes were observed in three males and three females. The mitotic metaphase chromosomes were observed in both sexes. The diploid chromosome number was 36 (
Fig. 1). The karyotypes were arranged by shape and size (
Fig. 5), and
Table 1 shows the results of chromosome measurement from this species. Chromosomes in males and females ranged from 2.2 to 4.2 µm in length. Mean total length of the diploid complements were 49.5±2.72 µm. Karyotype consists of nine pairs of metacentrics and nine pairs of submetacentric chromosomes (
Fig. 5).
Semisulcospira dolichostoma
Thirty-four chromosomes were observed at spermatogonial metaphase in 24 mitotic cells studied. The mean total chromosome length, based on measurements from three sets of karyotyped cells, was 50.3±2.29 µm. The length of chromosomes ranged from 2.1 to 4.8 µm (
Table 2).
Fig. 6 shows the karyotype constructed from the chromosomes shown in
Fig. 2, which was one of the most elongated complements. This species has 34 chromosomes: three pairs of metacentrics and 14 pairs of submetacentrics.
Viviparus rivularis
From three male specimens, spermatogonial mitotic divisions were observed in 18 cells. In these cells, 64 chromosomes (2n) were also counted (
Fig. 3). Meiotic chromosomes in male were observed (
Fig. 4). The lengths of the chromosomes in male ranged from 1.6 to 4.6 µm. Mean total length of the diploid complements in male were 96.4±7.86 µm (
Table 3). Karyotypes consist of two pairs of metacentrics and 30 pairs of submetacentric chromosomes (
Fig. 7).
DISCUSSION
One of the most commonly encountered genera of freshwater snails in the Far East Asia is
Semisulcospira. More than 18 species and subspecies of this genus have been described from China (
Yen, 1939). Identification of these species is difficult due to the great amount of variation in shell shape, structure and coloration. In the Pleuroceridae, the family to which
Semisulcospira is presently referred about chromosomes, 12 species have been studied from Japan and eight species from Korea (
Patterson, 1967;
Burch, 1968;
Park, 1994).
Semisulcospira libertina (=
bensoni) is regarded as the most important intermediate host of
Paragonimus westermani (
Yokogawa et al., 1960a,
1960b). Several nominal subspecies of
S. libertina have also been implicated as intermediate hosts for paragonimiasis and metagonimiasis in humans and other mammals in the Far East. However, a little is known concerning the biological relationships of the nominal species of
Semisulcospira over its wide range of geographic distribution (China, Formosa, Japan and Korea), and the relationship of
S. libertina to other melaniid snails.
Semisulcospira dolichostoma is a pleuracerid species in mainland China, and no information on the susceptibility of human-infecting trematodes is available. Although some viviparid snails such as
Cipangopaludina spp. act as the second intermediate host of echinostome trematods, the parasitological role of
V. rivularis has not been reported yet. Therefore, further studies on the susceptibility tests of these snails to trematodes are needed. Patterson (
1967) found 18 bivalents in each of the specimens of
S. libertina that were studied from four localities in Japan. Inaba and Tanaka (
1953) reported
S. libertina to have eight pairs of chromosomes (n=8, 2n=16). In this study, however, all specimens of this species had 18 bivalents. Burch (
1968) also found
S. libertina to have a haploid chromo-some number of n=18. We therefore mention that Inaba and Tanaka's report (
1953) of only eight pairs of chromosomes for this species is in error. Burch (
1968) extended our cytological knowledge of
Semisulcospira by studying eight additional species in which the haploid chromosome numbers ranged from n=8 to n=20. With such a wide variation in chromosome numbers, the pleuroceridae has been proven to be a group of considerable cytogenetic interest. Park (
1994) has reported on the chromosomes of eight species of pleuroceridae in Korea. The six species of
Semisulcospira in his study had chromosome number of n=18 and 2n=36. It is obvious that chromosome number is more conserved in species of
Semisulcospira in Korea than those in Japan or China. Park (
1994) reported that the karyotyping is especially helpful for analyzing
Semisulcospira in Korea, since all species studied so far from Korea have the same chromosome number (n=18, 2n=36).
Semisulcospira forticosta and
S. tegulata can be distinguished by their karyotypes.
In the Viviparidae, the chromosome number of all species studied so far is n=7 to n=14 (
Patterson, 1969;
Zhou et al., 1988;
Park et al., 1997). In the most primitive viviparid subfamily, Bellamyinae, the haploid chromosome numbers of 11 species studied range from 8 to 11 (
Ramamoorthy, 1958;
Inaba, 1965;
Zhou et al., 1988). In the subfamily Viviparinae, the haploid number of eight species ranges from n=7 to n=13 (
Rainer, 1963). Recently, Park et al. (
1997) reported the chromosome numbers in two species of echinostome vector snails of the family Viviparidae in Korea. In the present study, however, the chromosome number of
V. rivularis is different from those of the former reports.
According to the above comparison, it is suggested that S. libertina is more conserved than S. dolichostoma or V. rivularis. DNA sequence analysis of specific genes could be valuable for further classification of the relationship of vector snails in different localities.
Notes
-
This study was supported by a grant of the '97 Good Health Research and Development Project, Ministry of Health and Welfare, Republic of Korea.
References
- 1. Burch JB. Cytotaxonomy of some Japanese Semisulcospira (Streptoneura: Pleuroceridae). J Conchyl 1968;107:3-52.
- 2. Burch JB. Freshwater molluscs. Man-made lakes and human health, Stanley NF and Alpers MP. 1975, New York, USA. Academic Press; pp 311-321.
- 3. Dillon RT Jr. Karyotypic evolution in pleurocerid snails. II. Pleurocera, Goniobasis, and Juga. Malacologia 1991;33:339-344.
- 4. Inaba A. Cytotaxonomic studies of freshwater gastropods (I). Venus 1965;23:223-228.
- 5. Inaba A, Tanaka H. Studies on the chromosome numbers of some freshwater gastropods. J Sci Hiroshima Univ Ser B div 1 1953;14:213-220.
- 6. Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas 1964;52:201-220.
- 7. Nakano D, Wada KT, Izawa K. Karyotype of the freshwater snail, Semisulcospira reiniana (Prosobranchia: Pleuroceridae). Venus 1994;53:43-47.
- 8. Park GM. Cytotaxonomic studies of freshwater gastropods in Korea. Malacol Review 1994;27:23-41.
- 9. Park GM, Jung Y, Chung PR. Cytological studies of two species of genus Cipangopaludina (Mesogastropoda: Viviparidae) in Korea. Korean J Malacol 1997;13:21-27. (in Korean).
- 10. Patterson CM. Chromosome numbers of some Japanese freshwater snails. Venus 1967;25:69-72.
- 11. Patterson CM. Chromosomes of molluscs. Proc Symposium on Mollusca. Part II. 1969, Marine Biology Association India (1968); pp 635-686.
- 12. Rainer M. Vergleichende chromosomen-messungen an Viviparus-Arten (Prosobranchia). 23. Jahresb Schweizer Ges f Vererb-Forschg S S G. Arch. d Julius Klaus-St ft f Vererb-Forschung 1963;38:61-68.
- 13. Ramamoorthy K. Chromosomes of Viviparus dissimilis (Muller) and Viviparus bengalensis (Lamarck) (Prosobranchia-Gastropoda). J Zool Soc India 1958;10:33-38.
- 14. Yen TC. Die Chinesischen Land-und Subwasser-Gastropoden des Natur-Museums Senckenberg. 1939, Alle rechte vorbehalten Germany; pp 1-234.
- 15. Yokogawa S, Cort WW, Yokogawa M. Paragonimus and paragonimiasis. Exp Parasitol 1960a;10:81-137.
- 16. Yokogawa S, Cort WW, Yokogawa M. Paragonimus and paragonimiasis. Exp Parasitol 1960b;10:139-205.
- 17. Zhou T, Zhou M, Wu ZD. Karyotypes of five species in the family Viviparidae. Acta Zool Sinica 1988;34:364-370.
Figs. 1-4
Meiotic and mitotic chromosomes of three Chinese snail species.
Fig. 1. Diploid chromosomes in metaphase I of Semisulcospira libertina. Fig. 2. Diploid chromosomes in metaphase I of Semisulcospira dolichostoma. Fig. 3. Diploid chromosomes in metaphase I of Viviparus rivularis. Fig. 4. Haploid chromosomes of V. rivularis.
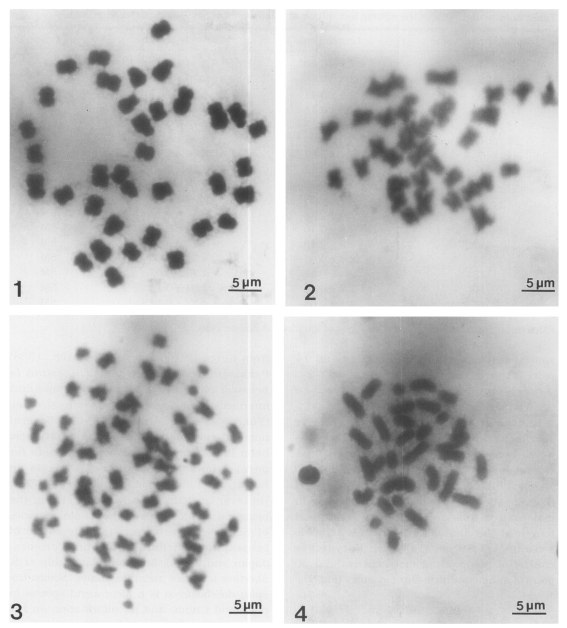
Figs. 5-7
Karyotypes of three Chinese snail species.
Fig. 5. Karyotype of the chromosomes in
Fig. 1 (
Semisulcospira libertina).
Fig. 6. Karyotype of the chromosomes in
Fig. 2 (
Semisulcospira dolichostoma).
Fig. 7. Karyotype of the chromosomes in
Fig. 3 (
Viviparus rivularis).

Table 1.Measurements of the mitotic metaphase chromosomes of
Semisulcospira libertinaa)
Table 1.
|
No. of chromosome |
Length (μm) |
Relative length |
Type |
|
1 |
4.1 ± 0.08 |
8.28 ± 0.22 |
Mb)
|
|
2 |
2.7 ± 0.31 |
5.45 ± 0.44 |
M |
|
3 |
2.5 ± 0.10 |
5.05 ± 0.05 |
M |
|
4 |
2.5 ± 0.08 |
5.04 ± 0.17 |
M |
|
5 |
2.3 ± 0.22 |
4.65 ± 0.46 |
M |
|
6 |
2.3 ± 0.08 |
4.64 ± 0.23 |
M |
|
7 |
2.2 ± 0.11 |
4.44 ± 0.44 |
M |
|
8 |
2.2 ± 0.06 |
4.44 ± 0.26 |
M |
|
9 |
2.2 ± 0.01 |
4.44 ± 0.08 |
M |
|
10 |
4.2 ± 0.21 |
8.48 ± 0.40 |
SMc)
|
|
11 |
3.3 ± 0.26 |
6.67 ± 0.50 |
SM |
|
12 |
3.2 ± 0.31 |
6.46 ± 0.47 |
SM |
|
13 |
3.2 ± 0.17 |
6.46 ± 0.12 |
SM |
|
14 |
2.7 ± 0.31 |
5.45 ± 0.34 |
SM |
|
15 |
2.7 ± 0.14 |
5.45 ± 0.10 |
SM |
|
16 |
2.5 ± 0.10 |
5.05 ± 0.05 |
SM |
|
17 |
2.4 ± 0.05 |
4.85 ± 0.01 |
SM |
|
18 |
2.3 ± 0.12 |
4.65 ± 0.23 |
SM |
Table 2.Measurements of the mitotic metaphase chromosomes of
Semisulcospira dolichostomaa)
Table 2.
|
No. of chromosome |
Length (μm) |
Relative length |
Type |
|
1 |
4.8 ± 0.10 |
9.54 ± 0.27 |
M |
|
2 |
3.4 ± 0.06 |
6.76 ± 0.27 |
M |
|
3 |
2.5 ± 0.44 |
4.97 ± 0.17 |
M |
|
4 |
3.8 ± 0.14 |
7.55 ± 0.46 |
SM |
|
5 |
3.6 ± 0.13 |
7.16 ± 0.05 |
SM |
|
6 |
3.5 ± 0.05 |
6.96 ± 0.25 |
SM |
|
7 |
3.4 ± 0.02 |
6.76 ± 0.44 |
SM |
|
8 |
3.1 ± 0.12 |
6.16 ± 0.30 |
SM |
|
9 |
2.9 ± 0.08 |
5.76 ± 0.54 |
SM |
|
10 |
2.7 ± 0.36 |
5.37 ± 0.19 |
SM |
|
11 |
2.6 ± 0.22 |
5.17 ± 0.20 |
SM |
|
12 |
2.5 ± 0.16 |
4.97 ± 0.17 |
SM |
|
13 |
2.4 ± 0.18 |
4.77 ± 0.13 |
SM |
|
14 |
2.4 ± 0.01 |
4.77 ± 0.06 |
SM |
|
15 |
2.4 ± 0.12 |
4.77 ± 0.03 |
SM |
|
16 |
2.2 ± 0.06 |
4.37 ± 0.15 |
SM |
|
17 |
2.1 ± 0.04 |
4.17 ± 0.14 |
SM |
Table 3.Measurements of the mitotic metaphase chromosomes of
Viviparus rivularisa)
Table 3.
|
No. of chromosome |
Length (μm) |
Relative length |
Type |
|
1 |
3.6 ± 0.14 |
3.73 ± 0.44 |
M |
|
2 |
2.1 ± 0.02 |
2.18 ± 0.42 |
M |
|
3 |
4.6 ± 0.26 |
4.77 ± 0.18 |
SM |
|
4 |
4.0 ± 0.42 |
4.15 ± 0.37 |
SM |
|
5 |
3.9 ± 0.13 |
4.05 ± 0.04 |
SM |
|
6 |
3.8 ± 0.11 |
3.94 ± 0.19 |
SM |
|
7 |
3.8 ± 0.08 |
3.94 ± 0.15 |
SM |
|
8 |
3.7 ± 0.15 |
3.84 ± 0.17 |
SM |
|
9 |
3.6 ± 0.14 |
3.73 ± 0.44 |
SM |
|
10 |
3.5 ± 0.21 |
3.63 ± 0.07 |
SM |
|
11 |
3.4 ± 0.11 |
3.53 ± 0.27 |
SM |
|
12 |
3.4 ± 0.14 |
3.52 ± 0.25 |
SM |
|
13 |
3.3 ± 0.12 |
3.42 ± 0.23 |
SM |
|
14 |
3.3 ± 0.07 |
3.42 ± 0.13 |
SM |
|
15 |
3.3 ± 0.10 |
3.42 ± 0.11 |
SM |
|
16 |
3.2 ± 0.22 |
3.32 ± 0.15 |
SM |
|
17 |
3.2 ± 0.14 |
3.32 ± 0.12 |
SM |
|
18 |
3.1 ± 0.08 |
3.22 ± 0.16 |
SM |
|
19 |
3.1 ± 0.04 |
3.22 ± 0.13 |
SM |
|
20 |
3.0 ± 0.22 |
3.11 ± 0.11 |
SM |
|
21 |
2.9 ± 0.25 |
3.00 ± 0.08 |
SM |
|
22 |
2.8 ± 0.06 |
2.90 ± 0.04 |
SM |
|
23 |
2.7 ± 0.17 |
2.80 ± 0.03 |
SM |
|
24 |
2.6 ± 0.28 |
2.70 ± 0.09 |
SM |
|
25 |
2.6 ± 0.11 |
2.70 ± 0.01 |
SM |
|
26 |
2.4 ± 0.31 |
2.49 ± 0.24 |
SM |
|
27 |
2.3 ± 0.04 |
2.39 ± 0.10 |
SM |
|
28 |
2.1 ± 0.02 |
2.18 ± 0.21 |
SM |
|
29 |
2.0 ± 0.15 |
2.07 ± 0.04 |
SM |
|
30 |
1.8 ± 0.04 |
1.87 ± 0.21 |
SM |
|
31 |
1.7 ± 0.28 |
1.76 ± 0.34 |
SM |
|
32 |
1.6 ± 0.14 |
1.66 ± 0.15 |
SM |
Citations
Citations to this article as recorded by

- An adaptable chromosome preparation methodology for use in invertebrate research organisms
Longhua Guo, Alice Accorsi, Shuonan He, Carlos Guerrero-Hernández, Shamilene Sivagnanam, Sean McKinney, Matthew Gibson, Alejandro Sánchez Alvarado
BMC Biology.2018;[Epub] CrossRef - Blood glycemia-modulating effects of melanian snail protein hydrolysates in mice with type II diabetes
Jae-Suk Choi, Joo-Wan Kim, Jeong Been Park, Sang Eun Pyo, Yong-Ki Hong, Sae Kwang Ku, Mi-Ryung Kim
International Journal of Molecular Medicine.2017; 39(6): 1437. CrossRef - Karyotype of Arion vulgaris Moquin-Tandon, 1856 (Gastropoda: Pulmonata: Arionidae)
Alexandr V. Garbar, Natalia S. Kadlubovska
Folia Malacologica.2014;[Epub] CrossRef